-
Notifications
You must be signed in to change notification settings - Fork 24
Population Loading
MABE allows users to load populations of organisms. These organisms are generated by MABE and stored in files named either P_organisms.csv or P_organisms_#.csv, where P is some arbitrary prefix chosen by the user and # is the a numeric value. Every organism generated by MABE has a unique attribute ID, and another attribute that contains the genome that encodes the organism. In addition, there may be additional attributes associated with this organism. Also, MABE may generate additional files that contain more attributes for the organisms, but you don't have to worry about it, MABE will figure that out.
The population to be loaded is specified in the parameter GLOBAL-initPop. This can be specified by writing a population loader script (with a .plf extension), e.g.
% GLOBAL-initPop = population_loader.plf
or directly as a single line of .plf syntax, e.g.
% GLOBAL-initPop = MASTER = 'LOD_organisms.csv'
All variables in a .plf script are collections of populations (think of them like 2D arrays). There is a special variable called MASTER, which is used by MABE to generate the initial population. MASTER must be a collection of exactly one population. The script can contain comments; anything following a # is a comment and is ignored. To illustrate .plf script syntax, consider the following directory layout |--mabe |--LOD_data.csv |--MyData |--LOD_data.csv
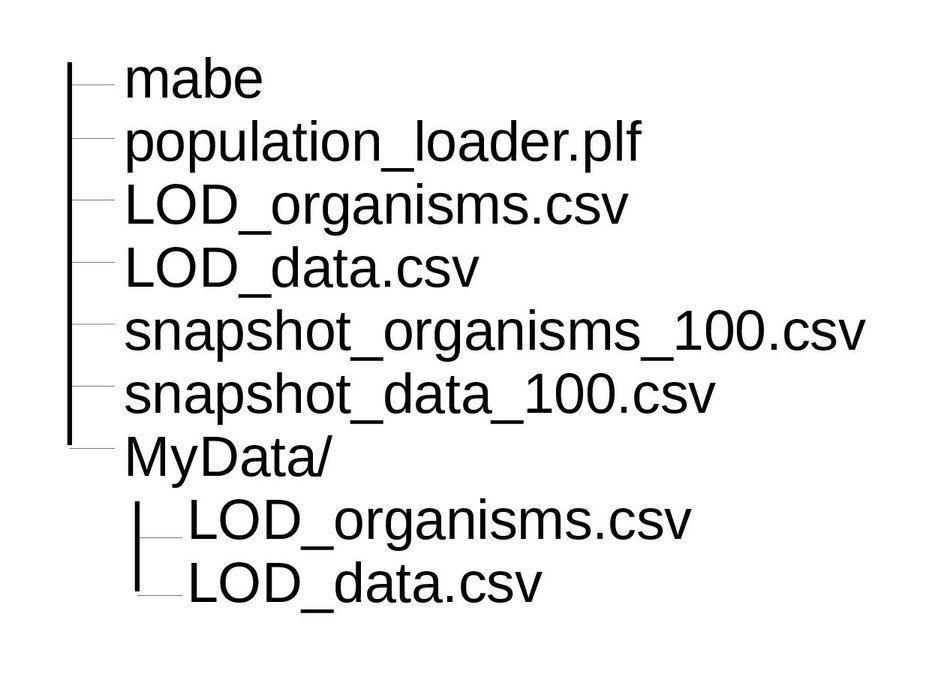
All organisms from a single file can be loaded simply by saying
MASTER = 'LOD_organisms.csv'
Note: A single file is treated as a single population.
Multiple populations/collections can be appended to each other. To do this, separate the populations/collections by : and enclose all of them in braces { }. e.g.
var = { 'snapshot_organisms.csv' : 'LOD_organisms.csv' }
File names to be loaded can be wildcarded. In addition, if the expanded wildcard matches multiple valid files, then all the files are appended to each other, creating a collection of files(populations). Hence, the line
var = { 'snapshot_organisms.csv' : 'LOD_organisms.csv' : 'MyData/LOD_organisms.csv' }
is equivalent to the following line
var = { 'LOD*.csv' : 'snapshot*.csv' : 'Mydata/*.csv' }
or even the following line
var = { '*' : 'Mydata/*' }
To collapse a collection with multiple populations into a collection with a single population, use the keyword collapse, e.g.
var = collapse { 'snapshot_organisms.csv' : 'LOD_organisms.csv' : 'MyData/LOD_organisms.csv' }
Populations can be specified more precisely by filtering the collections according to attributes. When a filter is applied to a collection, the filter is applied to each population within that collection, and the results are maintained in the same populations. The available filter keywords are
- greatest
This ranks the organisms according to the numeric values of the specified attribute, and selects the specified number of organisms with the greatest rank, from each population in the collection, e.g.
var = greatest 10 by RedSpots from '*' # var is now a collection of 2 populations, each with 10 organisms
- least
This ranks the organisms according to the numeric values of the specified attribute, and selects the specified number of organisms with the least rank, from each population in the collection, e.g.
var = least 5 by TailLength from '*' # var is now a collection of 2 populations, each with 5 organisms
- any
This randomly picks the specified number of organisms from each population in the collection e.g.
var = any 20 from '*' # var is now a collection of 2 populations, each with 20 organisms
- match
This picks only those organisms from each population in the collection, that exactly match the specified attribute e.g. To load organisms from all files in the current directory recursive, only if they have the ID 1729, you can say
MASTER = match ID where 1729 from { '*' : '*/*' } # MASTER now has at most 3 organisms
Note: match does an exact string comparison, so if the attribute has a value "6.30" for an organism, and you try to match "6.3", this attribute will not count as a match.
- <number>* (duplicate copies)
This copies every organism in each population in the collection, for the specified number of times, e.g.
var = greatest 10 by RedSpots from '*' var_plus = 30 * var # var_plus is now a collection of 2 populations, each with 300 organisms
The default keyword generates an organism according to the genome/brain type specified in the settings files. In fact, this is a common use case, where MABE is run with a default population of say, 100 organisms. The syntax to do that would simply be,
MASTER = default 100
You can use variables to store collections, or you can write them directly, as suits your preference (clear variable naming is usually helpful for other people reading your scripts). The following script,
best = greatest 5 by BlueSpots from '*'
worst = least 5 by GreenSpots from '*'
all = collapse { 10 * best : 5 * worst : default 50 }
MASTER = any 100 from all
is equivalent to the following one-liner,
MASTER = any 100 from {
collapse {
10 * { greatest 5 by BlueSpots from '*' }
: 5 * { least 5 by GreenSpots from '*' }
: default 50
}
}
Although one version may be more readable than the other
MASTER must be a collection with exactly one population. The following line is an error,
MASTER = '*' # error: MASTER is a collection of 2 populations
If a filter is applied on a collection, that filter must be applicable to every population in the collection.
In particular, if a filter specifies a number of organisms to be filtered, then every population in the collection must contain at least those many organisms.
var = least 5 by ID from { greatest 1 by ID from '*' } # error: least doesn't have enough organisms to pick 5 from each population.
If a filter specifies a certain attribute, then every organism in every population in the collection must have that attribute.
var = least 10 by Height from { '*' : default 100 } # error: default organisms don't have an attribute "Height"
home
welcome
MABE Parameter Widget
Installation and quick start
license
citations
release notes
developer contributions
consistency testing
Using MABE
Using Settings Files
Output Files
Creating Graphs with python
MABE framework
Defining Update
Brains
Markov Brain
Neuron Gate
Wire Brain
Human Brain
ConstantValues Brain
CGP Brain
Genetic Programing Brain
Artificial Neural Networks
Brains Structure and Connectome
Genomes
Circular Genome
Multi Genome
Genome Handlers
Genome Value Conversions
Organisms
Groups
Archivists
popFileColumns
Optimizers
Lexicase Optimizer
Worlds
Berry World
ComplexiPhi World
MultiThreadTemplate World
Utilities
DataMap
Parameters
Parameters Name Space
Adding Parameters to Code
ParametersTable
MTree
sequence function
Population Loading
PythonTools
MBuild
MGraph
MQ
findRelatedness
generatePhylogeny
Information Theory Tools
Brain States and Life Times
TimeSeries
Entropy Functions
Smearing
Fragmentation
State to State
Brain Infomation Tools
ProcessingTools