-
Notifications
You must be signed in to change notification settings - Fork 24
Home

Click here for installation and quick start instructions
Click here for a detailed breakdown of MABEs parts
MABE creates and manages populations of evolving digital organisms which are evaluated in worlds. Using the results of the evaluations new populations are generated by means of natural, artificial, and/or sexual selection.
The purpose of MABE is twofold:
- To support academic research into topics related to evolution:
- genetic dynamics
- haploid vs. diploid
- behavior
- intelligence
- sex and sexual selection
- To provide insight into evolution and evolutionary processes
To support the second aim, MABE encourages users to document parameters as they add them. As a result, MABE auto generates configuration files with usage messages. This allows a user to start experimenting with MABE without needing to do significant setup.
MABE avoids complex programming methods. Where we have employed more advanced methods it has been either to make the code easier to work with or significantly faster. In addition, these more advanced methods are used only in low-level features which most users are unlikely to directly encounter.
MABE has been designed to run on Windows, Mac, and Linux. MABE is written in c++11 using only standard libraries.
Before getting into Details it is useful to get a broad idea of the parts of MABE.
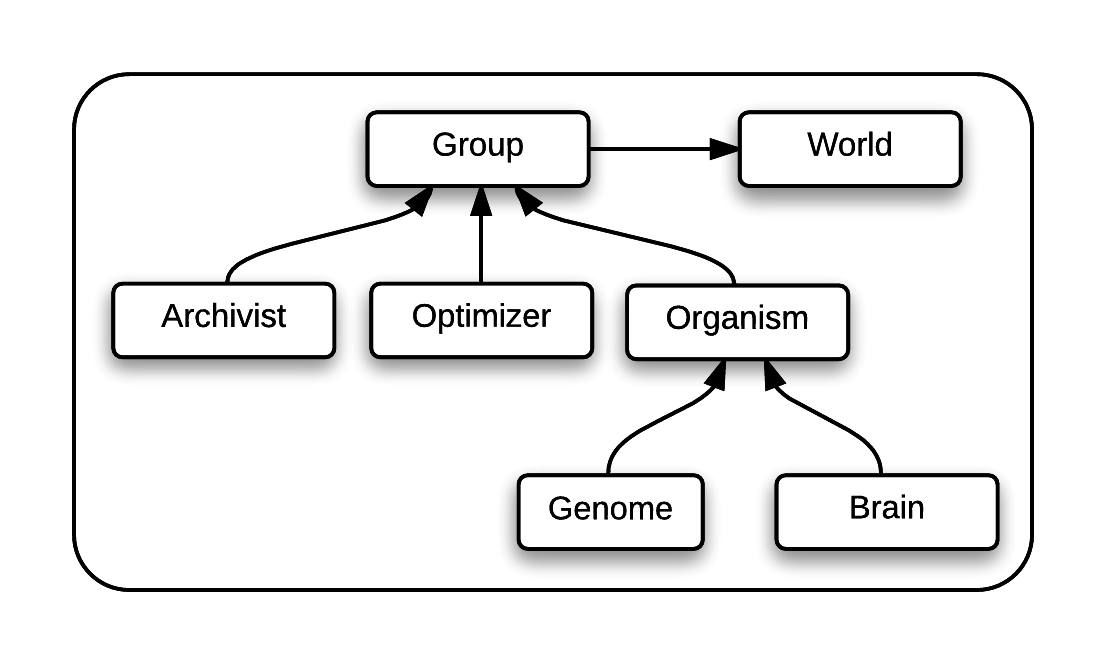
Let us consider an experiment involving lab rats being tested in a maze.
The rat (an Organism) contains a brain which determines how the rat interacts with the maze, and a genome, which provides a blueprint for the brain. The digital test subjects in MABE are called Organisms and the rat brain and genome are synonymous with the brain and genome in MABE.
A collection of rats makes up a population. In MABE, a collection of Organisms is also called a population.
The maze in this experiment is synonymous with the world in MABE.
MABE's archivist is synonymous with a person who collates all the data collected by the various scientists on the experiment and decides what data needs to be stored.
MABE's optimizer is synonymous with a person who decides which rats will be mated to produce the next generation.
A group in MABE is made up of a population of organisms, an optimizer and an archivist.
MABE development is lead by Clifford Bohm. Click here to visit his page.
home
welcome
MABE Parameter Widget
Installation and quick start
license
citations
release notes
developer contributions
consistency testing
Using MABE
Using Settings Files
Output Files
Creating Graphs with python
MABE framework
Defining Update
Brains
Markov Brain
Neuron Gate
Wire Brain
Human Brain
ConstantValues Brain
CGP Brain
Genetic Programing Brain
Artificial Neural Networks
Brains Structure and Connectome
Genomes
Circular Genome
Multi Genome
Genome Handlers
Genome Value Conversions
Organisms
Groups
Archivists
popFileColumns
Optimizers
Lexicase Optimizer
Worlds
Berry World
ComplexiPhi World
MultiThreadTemplate World
Utilities
DataMap
Parameters
Parameters Name Space
Adding Parameters to Code
ParametersTable
MTree
sequence function
Population Loading
PythonTools
MBuild
MGraph
MQ
findRelatedness
generatePhylogeny
Information Theory Tools
Brain States and Life Times
TimeSeries
Entropy Functions
Smearing
Fragmentation
State to State
Brain Infomation Tools
ProcessingTools