Convert 100 MIMIC III patients to the OHDSI OMOP Common Data Model and consider the implications of doing so.
Conversion to OMOP involves two distinct activities: first, data should be structure in the architecture of the OMOP Common Data Model. Second, information in the tables must be represented in standard vocabularies. The core standard vocabularies in the OMOP system are:
- RxNorm for drugs and medications
- LOINC for measurements in the lab or other documents
- SNOMED-CT for virtually everything else
Building a complete CDM strictly from note text would be a significant struggle because foundation information, such as the patient's identity, should be firmly established before attaching extracted information to it.
In this document, I will describe a recommended approach of converting structured data from the MIMIC III dataset to OMOP and then generating NLP-Derived information to attach to it. The Extract/Transform/Load (ETL) process used for structured data was supplied in an MIT github repository. The core technical challenges surround correctly locating and recoding healthcare entities in text. The core business challenge is determining what of this information can be trusted and for what purposes.
This document will describe an overall approach, describe the OMOP CDM standard tables and the NLP-related aspects of each, and then a potential workflow that includes a Proof-of-Concept (POC) NLP pipeline that is a starting point.
At a very high level, the JSL models and pipelines are well-equipped to perform the information extraction tasks we desire. Overall, any customer will require validation of NLP results before accepting them in a dataset to be used for their individual needs. Each subject area comes with some challenges that are detailed further in the table-by-table discussion below.
The fundamental reason for a database structured in OMOP CDM is to produce data for observational research that can be pooled and shared with other institutions. There are other reasons to do it, of course. OMOP is a helpful meeting place when building a research data warehouse. Many cutting-edge algorithms are published over data in OMOP form and these products can be put into use easily if your data is already in this shape. Before we make any hard decisions here, we would need to talk to the customer to understand their reasons for the conversion and their goals.
Core tables such asCONDITION and MEDICATION are first and foremost intended
for defining cohorts and the consumers of this information must know the
provenance and trustworthiness of the data. NLP-derived information needs to be
a known quantity before adding it to core tables without an extensive
discussion.
Reasons for including NLP information include:
- adding patient conditions and information from a problem list stored as text
- identifying medications not stored in structured data
- adding information that is not present in the structured data (such as non-Alcohol Use Disorder levels of drinking)
- identifying other Social Determinants of Health
- (in the dentistry use case) identifying all non-dental diagnoses and conditions because they appear nowhere else
- locating information about care providers missing in structured data
- adding family history
There is a lively conversation about whether data derived from an NLP process
should be used for cohort selection or outcomes. The standing compromise is that
any information inserted via NLP should have an appropriate flag in the
source_concept_id field for the table. Valid IDs are:
| concept_id | concept_name | domain_id | vocabulary_id |
|---|---|---|---|
| 32858 | NLP | Type Concept | Type Concept |
| 32423 | NLP derived | Type Concept | Meas Type |
| 32445 | NLP derived | Type Concept | Observation Type |
| 32426 | NLP derived | Type Concept | Drug Type |
| 32425 | NLP derived | Type Concept | Procedure Type |
| 32424 | NLP derived | Type Concept | Condition Type |
The rules we select for what should and should not be migrated from NLP derived information must always go back to first principles: who wants this information, and what do they want it for?
- when does an NLP-derived value earn a place in the
NOTE_NLPtable?- I propose that nothing with confidence below 0.95 should come over. There's a marked spike at very high levels of confidence and no clear cutoff below that.
- when does an NLP-derived value earn a place in the data table? Pending a
customer's preferences, a good starting set of guidelines is:
- Only entities with an Assertion of
Presentmove - Entities move to the table associated with the mapped concept's Domain ID
- Entities that move to tables must have an appropriate "NLP-Derived"
source_concept_idassociated - Entities aimed at
observationrequire extra scrutiny for attribution to family, history, etc
- Only entities with an Assertion of
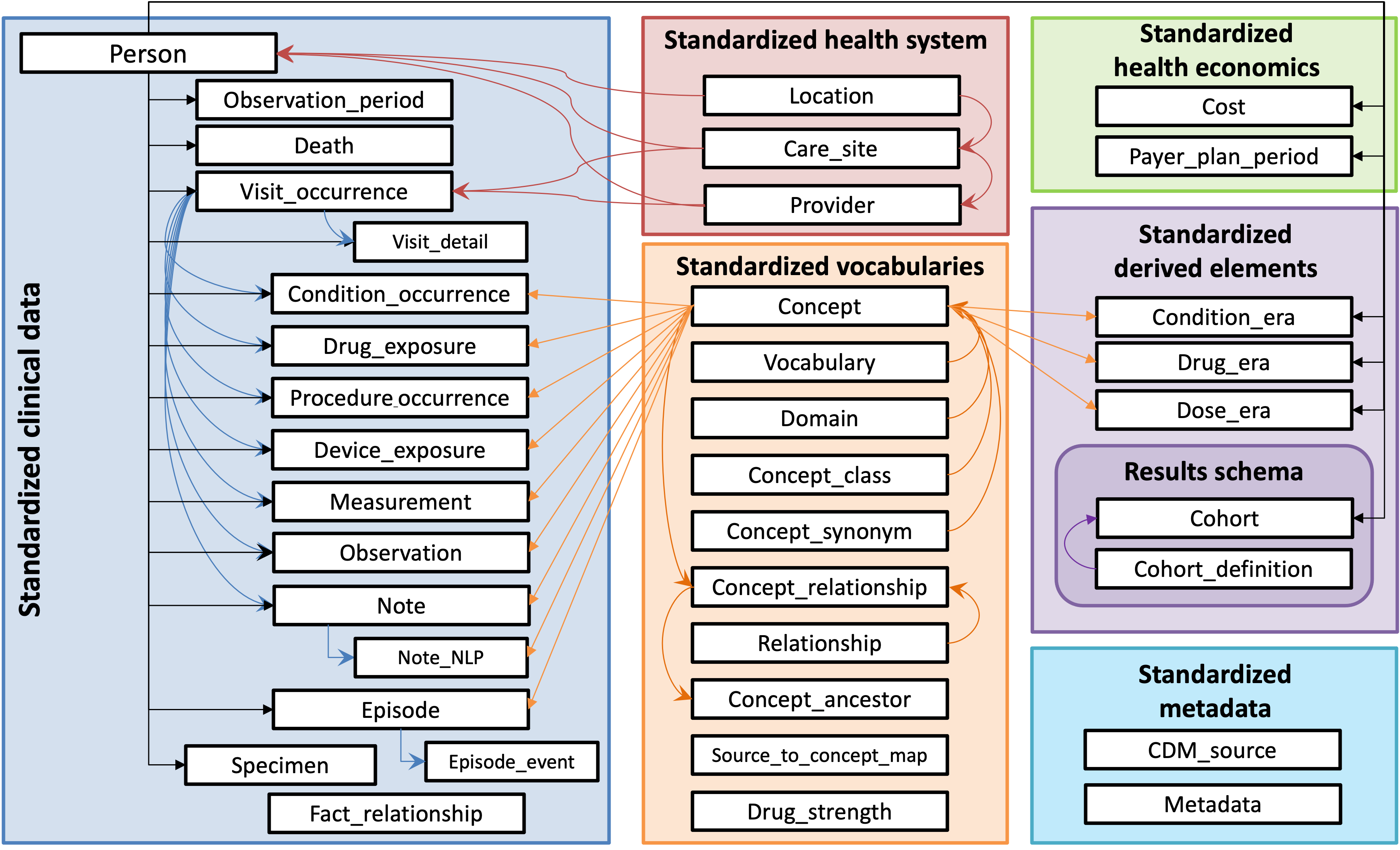
This section describes each of the core OMOP CDM tables and how we can approach them in the context of a conversion that includes NLP data.
person- information that should come solely from structured data. If our goal is to do a full conversion strictly from text, we can start to build out by using a JSL tool to extract key demographic data. There is real risk of entangling information about the patient's family in this information. A processing pipeline would have to include logic for rationalizing conflicting data.death- this is derived from theadmissiontable when a value is present indeathtime. This should be derived from structured data.visit_occurrence- derived fromadmission. If relying strictly on text data, some clients might accept that avisit_occurrencewas any contiguous block of days in which a note was written - but this is an imperfect assumption.visit_detail- a complex blend ofadmissionandtransfer, this describes a patient's specific encounters and transfers during a visit. The MIMIC III data does not bring over individualproviderinformation, which is a pity, and may be a where NLP could fill in some information. If relying solely on text data, I would not be inclined to populate this table.observation_period- mapped frommimiciii.admission. It's important to remember that MIMIC III is a dataset derived from ICU care: patients in a more general health care system are going to have different types of encounter and this mapping will become more complex. It should be derived from structured data.condition_occurrence- the MIMIC III ETL brings over both Conditions and Observations from thediagnoses_icdtable. An overwhelming majority of rows brought over did not map properly. (580,434/716,595): this is an ETL failure and needs to be fixed. Other data merged in comes fromadm_diag_cpt. NLP-derived information that clearly maps to theCONDITIONdomain is a candidate for insertion.- The POC pipeline identifies only gross "Condition" or "Observation" values and does not do any relation mapping to connect them to parts of the body, severity, or other text modifiers. This is an important block of information and should be resolved before anything is carried over.
drug_exposure- This is principally drawn from themimiciii.prescriptionstable. For NLP-derived information, the POC pipeline currently brings in only the name of a drug. We'd be better off adapting theDrugNormalizermodel demo-ed in notebook 14.0 to identify complete collections of dose/form/etc. related information and standardize it before passing off to an RxNorm concept identifier.procedure_occurrence- The MIMIC III ETL did a fair-to-middling job bringing structured data over fromprocedureevents_mv. Additional NLP-derived values from the domainProcedureare good candidates. As before, the POC pipeline needs to be improved to collect text modifiers to concepts before sending a richer chunk to code mapping.device_exposure- Thener_jslentity detection can labelMedical_Deviceentities, and SNOMED-CT supports devices.measurement- The POC pipeline has a commented-out LOINC leg. It quickly became clear that simply extracting results without doing the relationship extraction between tests, results, and dates was producing nothing but noise. This data can be better extracted with some variant of the JSLre_test_result_datepipeline that then feeds off to code that does very careful handling to LOINC. Unlike RxNorm, where we can assemble drug/dose/frequency etc. into a single code, this information is a bad fit forNOTE_NLP. I thinkNOTE_NLPwould have to take a record with the collection of test/result/unit etc. all stuffed intoterm_modifierswith their codes. ETL logic that transforms selected records to themeasurementtable would re-assemble concept IDs into a record.observation- Per OHDSI:
"The OBSERVATION table captures clinical facts about a Person obtained in the context of examination, questioning or a procedure. Any data that cannot be represented by any other domains, such as social and lifestyle facts, medical history, family history, etc. are recorded here"
There is a clear-cut case for bringing over high-confidence entities in the
domain Observation, although the concept_classes of Attribute and
Morph Abnormality are of limited help without better relation extraction
binding them to their related concepts.
The JSL Family Assertion is being applied liberally to concepts that really
appear to belong to the patient - and at least in the version of the pipeline in
the POC, I can't force obvious family hx into text that reflects in the output.
I need to learn more about properly deriving and applying Assertions and
Relations before really committing information that isn't a straight up
Observation as far as OMOP Standard Concepts are concerned.
-
notecomes over frommimiciii.noteevents. A required field callednote_class_concept_idis missing: According to CDM 5.4, it is "A Standard Concept Id representing the HL7 LOINC Document Type Vocabulary classification of the note." If the provenance of a Note is important information to the end consumer, the ETL will have to be amended to calculate and populate this column. -
note_nlpis a critical table for this work. Each row gets a specific concept extracted by an NLP process, along with modifiers and metadata describing the extraction process. In many OMOP communities, NLP-derived data stops here. In the migration we envision here, in which NLP-derived data is destined for the main clinical tables, we need to store enough information to support a final translation. (Primary amongst the information we've been working with - themeasurementtable is a complex collection of information about both a test and its results, and disentangling that information will require some ETL logic.) -
episodeis relatively new and designed for study of diseases that unfold over time. It and its companionepisode_eventare not populated. -
specimenis populated with nearly 40M entries for blood, urine, etc. samples, but 11M of them did not map to a specific concept. While more investigation is required, it may be possible to trace back to information in the NLP record
Care Site and Provider are included in the MIMIC III ETL, Location is not.
No data about physical address is available in the MIMIC III dataset. The JSL
models include a model in the Finance library that can extract addresses, but
it would be very difficult to know whether the presence of an address should be
attributed to a patient, a hospital location, or the site of a car crash. I am
inclined to leave this collection of information in the purview of a customer's
internal institutional knowledge.
If we are imputing provider from NLP results, a logical approach would be to
detect person entity information from note metadata and use this as a reference
framework for NLP code that identifies person names in the note itself. Logic
must separate patients from providers, and work to impute the identity of a
provider who is referred to only by title and surname. For example, it is
logical that in a note with "Susan Smith" listed as the provider in note
metadata, "Dr Smith" probably refers to that person. Without a firm context like
this, however, surnames alone are of limited value.
These tables were pulled from OHDSI's Athena: I selected common vocabularies I
work with all the time and stayed strictly with English-language resources. The
tables were built and indexed in a new schema adjacent to the MIMIC III data.
source_to_concept_map is an artifact of the structured data migration process
and is populated with hand mappings from source systems to the OMOP concepts.
These are calculated by the ETL process from structured data after it is loaded
and are used to specify a span of time when a condition or a drug or a specific
dose was present in the patient. It allows cohort selection to select patients
who meet a criterion any time in the span of a condition_era, etc,
transforming many point-in-time data to a single range for searching.
If we bring many concepts for conditions or drugs over to the data set after they are derived via NLP, these scripts should not be run until after the NLP transfer is complete.
This information did not exist in OMOP CDM at the time the ETLs were created. No cost/payment information exists in MIMIC III.
I pulled a current set of standard concepts from Athena, and then executed OMOP CDM build scripts to establish the vocabulary tables, load them, and index them.
ETL from MIMIC III is a (relatively) solved problem and the structured data and notes can be carried over with code such as that available at MIT's MIMIC-OMOP repo. That repo is at least six years old and has fallen out of sync with current versions of CDM. The ETL scripts needed to be tweaked to run at all. As noted above, some transforms were unsatisfactory: thorough testing and revision is required for any production use.
This is in the pipeline_experiments.ipynb Jupyter notebeook. It is an adaptation of demonstration pipelines in the JSL Jupyter Notebooks. I would like to refine it to ensure that information is calculated efficiently (and not repeatedly).
- I chose to use
ner-jslbut am concerned about properly filtering entity types - I used Routers to send true
DRUGinformation over to an RxNorm resolver, and just about everything else to a SNOMED-CT Observation resolver - Early experiments with
TESTs were unsatisfactory, as all kinds of acronyms were getting context-less entity matches. I want to handle test information more carefully with relationship matching between tests, values, and units. Something like the drug normalizer might be the best starting point. - We collect simple assertions about each entity as it's being located, and the confidence. This initial pass has produced results that need review.
The diagram below is a rough view of a draft of the pipeline, although it's been constantly tweaked since.
The pipeline is modeled heavily after a lot of the JSL demo code and I'm grateful for that help. It needs relationship extraction, handling of tests and results, and potentially more finely-grained entity -> code mapping.
Because we're only handling 100 patients' notes, I collect the notes in one SQL query and then, in batches, put them through the pipeline, extract fundamental concepts and attributes from the annotation process, and send them to my Postgres database in a temporary holding table. I would very much like to learn more about effectively streaming from database through the pipeline and back to the database without this constant load/unload cycle.
The output table should be thoroughly indexed before moving to the next step.
The migration analyst must perform extensive exploratory analysis of the results with the client to understand the distribution of information being extracted and its quality. Manual chart review may be required by clients to ensure that the extracted data is of high enough quality to move. The outcome of this assessment may be improvements to the NLP pipeline, or a post-NLP filter (or both)
Some annotations shouldn't even go to the note_nlp table at all. I think
anything with a low confidence value should not. Since note_nlp does make
allowances for negation and other assertions, though, if the client wants it,
entities that are negated or tied to family (etc) can be move to note_nlp.
This is a SQL query that could generate new rows for note_nlp:
SELECT e.note_id,
concept_name, -- we don't need this for the final transform but it's helpful for a sanity check
e.confidence, -- likewise, this doesn't come over but is helpful information while troubleshooting
e.chunks as snippet,
e.begin as offset,
concept_id as note_nlp_concept_id,
e.code as note_nlp_source_concept_id,
'JSL spark-nlp' as nlp_system,
'2024-06-25' as nlp_date,
case when e.assertion = 'Present' then 1 else 0 end as term_exists,
case when e.assertion = 'Past' then 'Past' else '' end as term_temporal,
e.assertion as term_modifiers,
concept_code as code,
domain_id,
c.vocabulary_id,
concept_class_id
from omop.concept c
inner join omop.TMP_EXTRACTED_CODES_BAK e
on e.code = c.concept_code and e.vocabulary_id = c.vocabulary_id
where c.domain_id in ('Measurement', 'Drug', 'Condition', 'Observation', 'Procedure')
and not concept_class_id in ('Attribute', 'Morph Abnormality')
and e.confidence >= 0.95
and invalid_reason is NULL;This has to be considered extremely carefully, working with the customer hand-in-hand the whole way. The risk of moving NLP data to standard tables can be viewed on two axes:
- What is the probability that we will put incorrect data in the table?
- What is the cost of putting incorrect data in the table?
This information may vary from table to table, and the decision about it may also vary.
The probability of error should be assessed first internally, and then with the client by hand-reviewing the inference results over a selection of random records. The cost of error is entirely in the decision space of the client. Noting the use of a wheelchair by a patient when it's really the patient's mother may be trivial in a dataset that will be used to study cancer. It will be a completely different matter if the customer is interested in studying falls.
Another approach to managing error would be to algorithmically compare NLP-derived information to structured data from the same visit and closely inspect records where they are contradictory. We hope NLP will add missing data - but if there is an outright conflict in information, we should investigate.
It is entirely possible to build out an OMOP CDM dataset using only the information in clinical notes, but the extracted data is more trustworthy and meaningful when blended with structured data during the ETL. Aside from the usual NLP risks of mis-attributing information and mis-identifying entities, the information gathered with JSL's entity detection, concept identification, and attribution models can populate the core tables such as
personcondition_occurrenceobservationdrug_exposuremeasurementprocedure_occurrence
In the absence of structured data, other key tables such as visit_occurrence
and provider would require carefully-documented logic to impute information
from note metadata.
Our recommended approach is to build the dataset out with known-good structured
data and then supplement it with information from clinical notes. All quality
NLP-derived information can be stored in the OMOP NOTE_NLP table. After that,
rules for what information in this table might migrate to the core clinical
concept tables must be developed with the client, bearing in mind their end
purpose.