- Authors
- Statement of Need
- Summary
- Installation instructions
- Additional Installation
- Data processing examples
- Citing CusToM
- Contributing
- Code of conduct
- License
- Acknowledgements
- Bibliography
Tags:
- Motion analysis
- Subject-specific
- Kinematics
- Dynamics
- Muscle forces
- Ground reaction forces prediction
- Antoine Muller (1)
- Charles Pontonnier (1,2)
- Pierre Puchaud (1,2,3)
- Georges Dumont (1)
- (1) Univ Rennes, CNRS, Inria, IRISA - UMR 6074, F-35000 Rennes, France
- (2) Centre de Recherche des Ecoles de Saint-Cyr Coëtquidan, 56380 Guer, France
- (3) Univ Rennes, M2S - EA 1274, 35170 Bruz, France
date: 2 July 2018
Inverse dynamics based musculoskeletal analysis aims at calculating biomechanical quantities to understand motion from joint kinematics to muscle forces. Generic models generally based on cadaveric templates are used as an input. These generic models are based on three layers modelisation. The first one is geometrical defining the polyarticulated rigid body system and the kinematic joints. The inertial layer defines mass, centers of mass position and inertia matrix of rigid body of the polyarticulated system. And, the muscle layer defines the muscle paths and force generation behaviours of muscles. This generic models are then calibrated through multiple calibration steps to be subject-specific base on motion capture data. Finally the subject-specific model is used to understand recorded motions of the subject.
However, musculoskeletal simulation requires high computational cost. Editing and assembling features to modify generic models. Subject-specific calibrations and multiple simulations are required to compute subject-specific quantities on recorded trials. And current available musculoskeletal softwares are heavy and requires expertise like SIMM (MusculoGraphics, Inc., Santa Rosa, CA), Anybody (Anybody Technology, Aalborg, Denmark) and OpenSim OpenSim (Simtk, Stanford, CA). Moreover, SIMM and Anybody are commercial softwares. OpenSim is open source and available at https://github.com/opensim-org. Core simulation elements are however difficult to handle and modify for research purposes, therefore we thought necessary to build our own musculoskeletal simulation code. The source code was developped with Matlab to allow many researchers to understand and contribute to the code.
Customizable Toolbox for Musculoskeletal simulation (CusToM) is a MATLAB toolbox aimed at performing inverse dynamics based musculoskeletal analyzes (Erdemir et al., 2007). This type of analysis is essential to access mechanical quantities of human motion in different fields such as clinics, ergonomics and sports. CusToM exhibits several features. It can generate a personalized musculoskeletal model, and can solve from motion capture data inverse kinematics, external forces estimation, inverse dynamics and muscle forces estimation problems as in various musculoskeletal simulation software (Damsgaard et al. 2006, Delp et al. 2007).
According to user choices, the musculoskeletal model generation is achieved thanks to libraries containing pre-registered models (Muller et al. 2015). These. These models consist of body parts osteoarticular models, set of markers or set of muscles to be combined together. From an anthropometric based model, the geometric, inertial and muscular parameters are calibrated to fit the size and mass of the subject to be analyzed (Muller et al. 2016) (Muller, Pontonnier, and Dumont 2017) (A. Muller et al. 2017). Then, from motion capture data (extracted from a c3d file thanks to (Barre and Armand 2014)), the inverse kinematics step computes joint coordinates trajectories against time (Lu and O’connor 1999). Then, joint torques are computed thanks to an inverse dynamics step (Featherstone, 2008). To this end, external forces applied to the subject have to be known. They may be directly extracted from experimental data (as platform forces) or be estimated from motion data by using the equations of motion in an optimization scheme (Fluit et al, 2008). Last, muscle forces are estimated. It consists in finding a repartition of muscle forces respecting the joint torques and representing the central nervous system strategy (Crowninshield 1978) (Antoine Muller et al. 2017) (Muller, Pontonnier, and Dumont 2018).
For a large set of musculoskeletal models and motion data, CusToM can easily performs all of the analyzes described above. CusToM has been created as a modular tool to let the user being as free and autonomous as possible. The osteoarticular models, set of markers and set of muscles are defined as bricks customizable and adaptable with each other. The design or the modification of a musculoskeletal model is simplified thanks to this modularity. Following the same idea, some methods are defined as adaptable bricks. Testing new cost functions in the optimization schemes, changing performance criteria or creating alternative motion analysis methods can be done in a relatively easy way.
A user interface has been developed to facilitate the data management and the model definition during a given study.
You need to create a copy on a local directory on your machine to use CusToM. Obtain a copy by downloading and unzipping the latest zip file or clone CusToM instead e.g. using: git clone https://github.com/anmuller/CusToM. You can place the CusToM folder anywhere on your machine.
After downloading the main folder and placing it in a relevant location, the installation only consists in running the Installation file. It checks if you have a compatible version of Matlab, if the needed toolboxes are installed. The function will also add the Functions folder of CusToM to your current path.
CusToM was implemented and tested with the Matlab 2018a version on Windows 10. Authors can not guarantee that the code could be run on previous versions. Here is a list of toolboxes which appear to be used in CusToM:
- Symbolic Matlab Toolbox
- Optimization Toolbox
- Parallel Computing Toolbox
- DSP System Toolbox or Signal Processing Toolbox
- Robotics System Toolbox
CusToM was not developped on MacosX and Linux. For MacosX, it could be necessary to download the source files of BTK and to compile and install BTK accordingly with your device. You would need to download btk-core-0.3.0_src.zip. For Linux, you would need to download [btk-0.3.0_Linux_MatlabR2009a_64bit.tar.gz] or [btk-0.3.0_Linux_MatlabR2009a_32bit.tar.gz] depending on your device.
For Ubuntu, the content of lib folder in btk must be moved in /usr/local/lib. For Fedora, the content of lib folder in btk must be moved in /usr/lib64. Then, use the command sudo ldconfig in the new location.
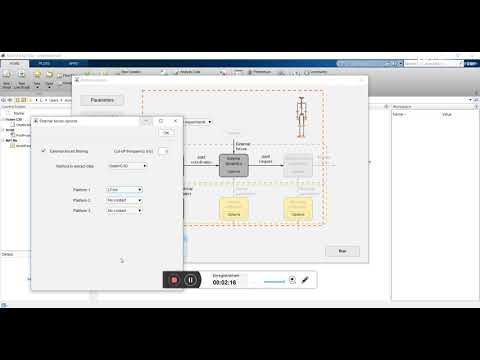
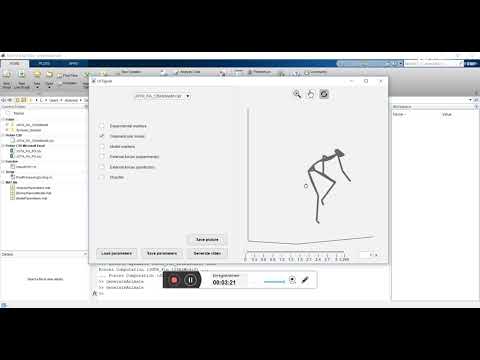
To check that your installation of btk is correct, go whitin any folder of "Examples". Write "GenerateParameters" in the Matlab command, click on "GenerateParameters" in the new window. Then, in the window "Analysis" which will open, desactivate all the boxes except the InverseKinematics box. Click on "Run", and check that there is no error in Matlab prompt due to unreadable .c3d file. If there is no error, the installation is complete. If not, feel free to open an issue in github.
-
BTK Biomechanical toolkit to import .c3d files
-
3D Animations tools have incorporated thanks to GIBBON which has an advanced graphics animation creation and exporting capabilities through a figure window based GUI.

-
Export_fig, it helps to export publication quality images.
- Full body model (source : AMMR)
- Twente lower extremity model 2.0
- Gait 2354 model
- Kinematic model of Xsens MVN
Four tutorials extracted from research works are available in the current release. The first one consists in predicting the ground reaction forces on a sidestep motion. The second tutorials consists in analyzing the kinematics of a pick-and-place task realized in a Virtual Reality Environment (holding a Head-Mounted-Display). The third tutorial consists in estimating the lower limbs muscle forces during a cycling motion. The tutorials are also illustrated by videos available in the repository. As the videos were recorded using a older version of CusToM, you may notice small differences in the user interface. You can either follow the instructions below or the videos to run these examples.
Examples are both detailed in the CusToM Documentation and in Youtube Videos.
A fourth tutorial has been added on a tennis service motion from XSENS Analyze suite based on inertial sensors.
The first example is a side-step motion. It is extracted from a database currently being developed for population characterization. The objective of the tutorial is to compare measured and predicted external forces.
The second example is a pick-and-place motion realized in a virtual environment with an haptic device. The task is similar to the ones analyzed in. Here since no direct measurement of the external forces was made, the study focused only on kinematics features.
This third example consisted in linking the symmetry, the performance and the health during a cycling motion. The muscular symmetry is analyzed by the motion analysis containing an inverse kinematics step, an inverse dynamics step and a muscle forces estimation step. The markers set used is that called Marker_set2 in CusToM. External forces applied on each foot were measured with two mobile platforms located on pedals.
The objective of the last tutorial is to evaluate ground reaction forces of a tennis service motion recorded with XSENS inertial sensors using the force prediction algorithm. At the end of the tutorial, key concepts should be understood: contact points, contribution to dynamic equilibrium.
Extra tutorials could be found on the workshop repository.
Cite the Journal of Open Source Software paper:
Muller, A., Pontonnier, C., Puchaud, P., Dumont, G., (2019). CusToM : a Matlab toolbox for musculoskeletal simulation, Journal of Open Source Software.
See CONTRIBUTING
See CODE_OF_CONDUCT
CusToM is provided under: . The file can be found on the repository.
We acknowledge contributions from Diane Haering, Félix Demore, Marvin Chauwin, Claire Livet, Lancelot Barthe and Amaury Dalla Monta.
Barre, A., and S. Armand. 2014. “Biomechanical Toolkit: Open-Source Framework to Visualize and Process Biomechanical Data.” Computer Methods and Pro- grams in Biomedicine 114 (1). Elsevier:80–87. https://doi.org/10.1016/j.cmpb.2014.01. 012.
Crowninshield, Roy D. 1978. “Use of Optimization Techniques to Predict Muscle Forces.” Journal of Biomechanical Engineering 100 (2). American Society of Mechanical Engineers:88–92. https://doi.org/10.1115/1.3426197.
Damsgaard, M., J. Rasmussen, S. T. Christensen, E. Surma, and M. de Zee. 2006. “Analysis of musculoskeletal systems in the AnyBody Modeling System.” Simulation Modelling Practice and Theory 14 (8):1100–1111. https://doi.org/10.1016/j.simpat.2006. 09.001.
Delp, S. L., F. C Anderson, A. S Arnold, P. Loan, A. Habib, C. T. John, E. Guendelman, and D. G. Thelen. 2007. “OpenSim: Open source to create and analyze dynamic simu- lations of movement.” IEEE Transactions on Bio-Medical Engineering 54 (11):1940–50. https://doi.org/10.1109/TBME.2007.901024.
Erdemir, A., S. McLean, W. Herzog, and A. J. van den Bogert. 2007. “Model-based estimation of muscle forces exerted during movements.” Clinical Biomechanics 22 (2):131– 54. https://doi.org/10.1016/j.clinbiomech.2006.09.005.
Featherstone, R., 2008. Rigid Body Dynamics Algorithms, Constraints.
Fluit, R., M. S. Andersen, S. Kolk, N. Verdonschot, and H. F. J. M. Koopman. 2014. “Pre- diction of ground reaction forces and moments during various activities of daily living.”Journal of Biomechanics 47 (10). Elsevier:2321–9. https://doi.org/10.1016/j.jbiomech. 2014.04.030.
Lu, T. W., and J. J. O’connor. 1999. “Bone position estimation from skin marker co- ordinates using global optimisation with joint constraints.” Journal of Biomechanics 32 (2):129–34. https://doi.org/10.1016/S0021-9290(98)00158-4.
Muller, A., C. Germain, C. Pontonnier, and G. Dumont. 2016. “A simple method to calibrate kinematical invariants: application to overhead throwing.” In Proceedings of the 33rd International Society of Biomechanics in Sports.
Muller, A., D. Haering, C. Pontonnier, and G. Dumont. 2017. “Non-invasive techniques for musculoskeletal model calibration.” In Proceedings of the 23ème Congrès Français de Mécanique.
Muller, A., C. Pontonnier, and G. Dumont. 2017. “Uncertainty propagation in multibody human model dynamics.” Multibody System Dynamics 40 (2):177–92. https://doi.org/10. 1007/s11044-017-9566-7.
Muller, A., C. Pontonnier, C. Germain, and G. Dumont. 2015. “Dealing with modularity of multibody models.” In Proceedings of the 40ème Congrès de La Société de Biomécanique, Computer Methods in Biomechanics and Biomedical Engineering, 18:2008–9. sup1. https://doi.org/10.1080/10255842.2015.1069599.
Muller, A., F. Demore, C. Pontonnier, and G. Dumont. 2017. “MusIC Makes the Muscles Work Together.” In XVI International Symposium on Computer Simulation in Biomechanics, 2.
Muller, A., C. Pontonnier, and G. Dumont. 2018. “The Music Method: A Fast and Quasi-Optimal Solution to the Muscle Forces Estimation Problem.” Computer Methods in Biomechanics and Biomedical Engineering 21 (2). Taylor & Francis:149–60. https://doi.org/10.1080/10255842.2018.1429596