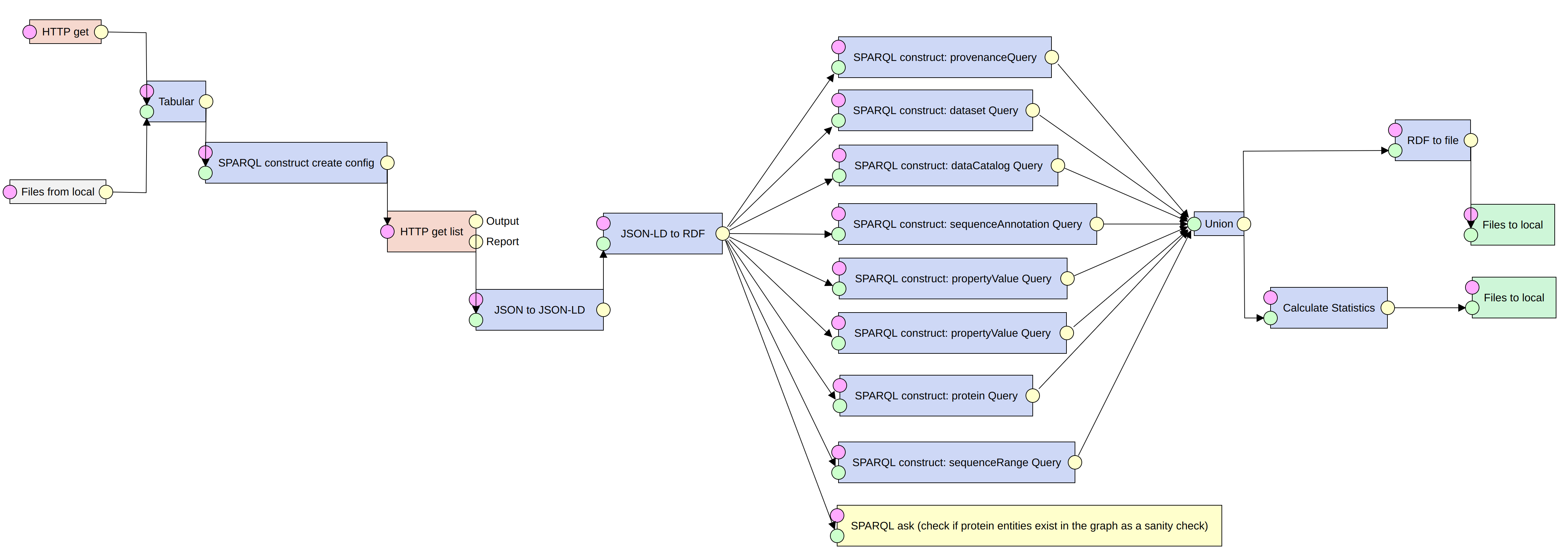
An ETL pipeline to construct the Intrinsically Disordered Proteins Knowledge Graph (IDP-KG) using Bioschemas JSON-LD data dumps
As part of the one-week Biohackathion Europe 2022 in Paris France, a group was formed to work on Project 23 titled, “Publishing and Consuming Schema.org DataFeeds.” Schema.org and Bioschemas, which is built on top of it (Gray et al., 2017), are lightweight vocabularies that aim at making the contents of web pages machine-readable so that software agents can consume that content and understand it in an actionable way. Due to the time needed to process each page, extracting markup by visiting each page of a site is not practical for huge sites. This approach imposes processing requirements on the publisher and the consumer. The Schema.org community proposed a method for exchanging markup from various pages as a DataFeed published at a recognized address in February 2022. The feed could consist of a single file containing the entire information or it could be divided into different files based on different aspects of the dataset, such as proteins and molecular entities, as in the case of ChEMBL. This would ease publisher and customer processing requirements and accelerate data collection. The aim of the project is to explore the implementation of the Schema.org proposal from both a producer and consumer perspective, for a variety of resources implementing different Bioschemas profiles. This report focuses on the consumer part of the project proposal where we explored an ETL pipeline (Extract-Transform-Load) approach and implemented a consumption pipeline that enables data dumps to be ingested into knowledge graphs (KG).
Preprint: https://biohackrxiv.org/7f95d/