Kd trees for Julia.
Note: This package is deprecated in favor of NearestNeighbors.jl which can be found at: https://github.com/KristofferC/NearestNeighbors.jl.
This package contains an optimized kd tree to perform k nearest neighbour searches and range searches.
The readme includes some usage examples, different benchmarks and a comparison for kNN to scipy's cKDTree.
The tree is created with:
KDTree(data [, leafsize=10, reorder=true])The data argument for the tree should be a matrix of floats of dimension (n_dim, n_points). The argument leafsize determines for what number of points the tree should stop splitting. The default value is leafsize = 10 which is a decent value. However, the optimal leafsize is dependent on the cost of the
distance function which is dependent on the dimension of the data.
The reorder argument is a bool which determines if the input data should
be reordered to optimize for memory access. Points that are likely to be accessed close in time are also put close in memory. The default is to enable this.
The function knn(tree, point, k) finds the k nearest neighbours to a given point. The function returns a tuple of two lists with the indices and the distances from the given points respectively. These are sorted in the order of smallest to largest distance.
using KDTrees
tree = KDTree(randn(3, 1000))
knn(tree, [0.0, 0.0, 0.0], 5)gives both the indices and distances:
([300,119,206,180,845],[0.052019,0.200885,0.220441,0.22447,0.235882])
The function inball(tree, point, radius [, sort=false]) finds all points closer than radius to point. The function
returns a list of the indices of the points in range. If sort is set to true, the indices will be sorted before being returned.
using KDTrees
tree = KDTree(randn(3, 1000))
inball(tree, [0.0, 0.0, 0.0], 0.4, true)gives the indices:
8-element Array{Int64,1}:
184
199
307
586
646
680
849
906
926
KDTrees.jl also supports dual tree range searches where the query points are put in their own separate tree and both trees are traversed at the same time while extracting the pairs of points that are in a given range.
Dual tree range searches are performed with the function inball(tree1, tree2, radius [, sort=false])
and returns a list of list such that the i:th list contains the indices for the
points in tree2 that are in range to point i in tree.
If sort = true the lists are sorted before being returned. Currently, trees where the
data has been optimized for memory allocation is not supported. This function has not
gotten the same amount of optimization as the others so it might be faster just to
loop through the points one by one.
using KDTrees
tree = KDTree(rand(1, 12), reorder = false)
tree2 = KDTree(rand(1, 16), reorder = false)
inball(tree, tree2, 0.1)gives the result
12-element Array{Array{Int64,1},1}:
[16,11,15,5,9,14]
[6]
[5,7]
[6]
[5,7]
[10,3,2]
[5,7]
[4,1]
[16,12,11,15,9,14]
[4,1]
[7,6]
[5,7]
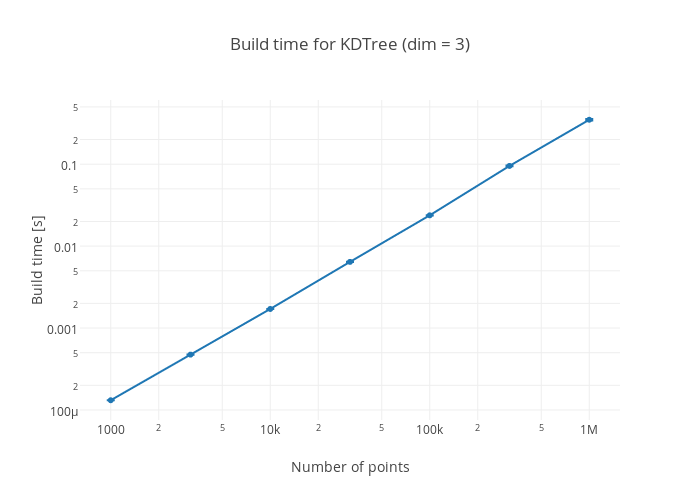
The benchmarks have been made with computer with a 4 core Intel i5-2500K @ 4.2
GHz with Julia v0.4.0-dev+3034 with reorder = true in the building of the trees.
Clicking on a plot takes you to the Plotly site for the plot where the exact data can be seen.
One of the most popular packages for scientific computing in Python is the scipy package. It can therefore be interesting to see how KDTrees.jl compares against scipy's cKDTree.
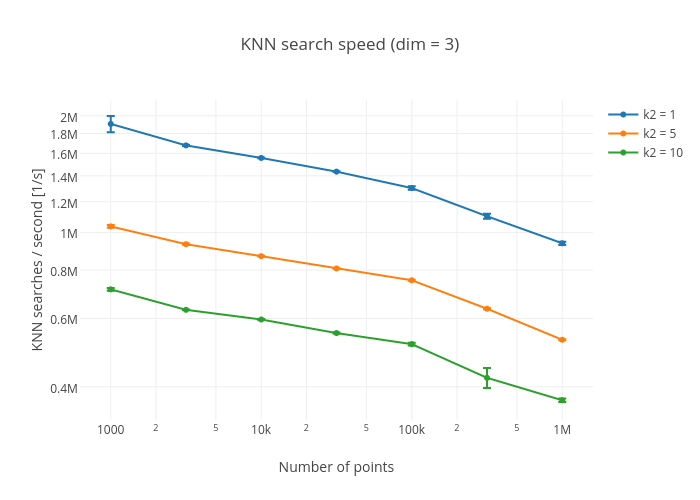
A KNN search for a 100 000 point tree was performed for the five closest neighbours. The code and the resulting search speed are shown, first for cKDTree and then for KDTrees.jl
cKDTree:
>>> import numpy as np
>>> from scipy.spatial import cKDTree
>>> import timeit
>>> t = timeit.Timer("tree.query(queries, k=5)",
"""
import numpy as np
from scipy.spatial import cKDTree
data = np.random.rand(10**5, 3)
tree = cKDTree(data)
queries = np.random.rand(10**5, 3)
""")
>>> t = min(t.repeat(3, 10)) / 10
>>> print("knn / sec: ", 10**5 / t)
('knn / sec: ', 251394)KDTrees.jl:
julia> tree = KDTree(rand(3,10^5));
julia> t = @elapsed for i = 1:10^5
knn(tree, rand(3), 5)
end;
julia> print("knn / sec: ", 10^5 / t)
knn / sec: 700675Contributions are more than welcome. If you have an idea that would make the tree have better performance or be more general please create a PR. Make sure you run the benchmarks before and after your changes.
Kristoffer Carlsson (@KristofferC)