-
Notifications
You must be signed in to change notification settings - Fork 24
create_weights
This file creates the weights file required to run frescalo, as outlined in (Hill, 2011). This function takes a table of geographical distances between sites and a table of numeric data from which to calculate similarity (for example, landcover or abiotic data).
This function requires you to have two tables, one a table of distances, and one an attributes table. sparta comes with an example of each. Here is the distance table:
# Load example distance table
data(dist)
head(dist)
Var1 Var2 value
1 SP00 SP00 0
2 SP01 SP00 10
3 SP02 SP00 20
4 SP03 SP00 30
5 SP04 SP00 40
6 SP05 SP00 50
This table has the names of the two sites between which we are measuring the distance in the first two columns and then the distance between them in the third column. If you are unsure how to make a table like this from your data look at the section below on how to make a distance table
The attributes table can be a table of abiotic factors or habitat types. In the example we have a table of habitat types, giving the proportion of each site covered by each habitat. The dist() function is then used within create_weights to calculate the distance between them. As a result it is important that your attributes are on a similar scale.
# Load example attributes table
data(sim)
head(sim)
SQ_SQUARE acidgrass1 arable10k bog10k bwood10k calcgrass1 cwood10k
SP00 SP00 0.00000000 0.5037063 0 0.05662500 0.00034375 0.00306875
SP01 SP01 0.00000000 0.4550000 0 0.14020625 0.00058125 0.00703125
SP02 SP02 0.00011250 0.3854250 0 0.10798125 0.00813750 0.00484375
SP03 SP03 0.00071875 0.5740688 0 0.06631875 0.00149375 0.00089375
SP04 SP04 0.00000000 0.5556250 0 0.03608125 0.00076875 0.00072500
SP05 SP05 0.00018125 0.5573000 0 0.07351250 0.00038125 0.00695625
dsheath10k fen_m_s10k freshwater heathergra imprgrass1 inlandrock
SP00 0.00000000 5.625e-05 0.00093125 0.00151250 0.3009938 0.00135625
SP01 0.00019375 0.000e+00 0.00033750 0.00261875 0.2486000 0.00099375
SP02 0.00000000 0.000e+00 0.00019375 0.00615000 0.2499500 0.00146250
SP03 0.00000000 0.000e+00 0.00058125 0.00246250 0.2319375 0.00261250
SP04 0.00000000 0.000e+00 0.00583125 0.00055000 0.2177625 0.00588750
SP05 0.00000000 0.000e+00 0.00223125 0.00028750 0.2600500 0.00740625
litt_rock1 litt_sed10 montane10k neutgrass1 roughgrass saltmarsh1
SP00 0 0 0 0.00000000 0.07934375 0
SP01 0 0 0 0.00000000 0.13689375 0
SP02 0 0 0 0.00342500 0.21804375 0
SP03 0 0 0 0.01321875 0.08954375 0
SP04 0 0 0 0.00483125 0.07753750 0
SP05 0 0 0 0.00712500 0.04665000 0
su_lit_roc su_lit_sed suburban10 urban10k
SP00 0 0 0.04326875 0.00879375
SP01 0 0 0.00689375 0.00065000
SP02 0 0 0.01370625 0.00056875
SP03 0 0 0.01465000 0.00150000
SP04 0 0 0.07882500 0.01557500
SP05 0 0 0.03086250 0.00705625
If your attributes are not on a similar scale you may want to normalise them. For example, you might have a measure of mean altitude in meters that ranges from 20m to 1000m and a measure of humidity that ranges from 0.6 to 0.9. In this scenario altitude will have a much greater impact on your similarity measure. To normalise your attributes you can use the arguement 'normalise = TRUE'. This divides all values in each column by the maximum for that column, re-scaling everything to 0-1. Since the example data is all measured as proportions I have chosen not to normalise. You can now run the weights creation function:
weights<-create_weights(dist=dist,
sim=sim,
dist_sub=20,
sim_sub=10)
Creating similarity distance table...Complete
Creating weights file...
0%
10%
20%
30%
40%
50%
60%
70%
80%
90%
100%
Complete
Creating the weights file can take a while since the function has to compare every site to every other site. To help see how things are going the function reports on progress to the console. The weights file produced is now in the format required by frescalo:
head(weights)
target neighbour weight
1 SP99 SP77 0.0084
2 SP99 SP78 0.0009
3 SP99 SP85 0.0013
4 SP99 SP87 0.0995
5 SP99 SP88 0.2889
6 SP99 SP89 0.0670
We can now use this weights file in frescalo as follows:
# Set up a directory to save output
sinkdir <- getwd()
# Load example dataset
data(ex_dat)
# Run frescalo
fres_out<-frescalo(Data=ex_dat,
sinkdir=sinkdir,
time_periods=data.frame(start=c(1980,1990),end=c(1989,1999)),
site_col='hectad',
sp_col='CONCEPT',
start_col='TO_STARTDATE',
end_col='Date',
Fres_weights=weights)
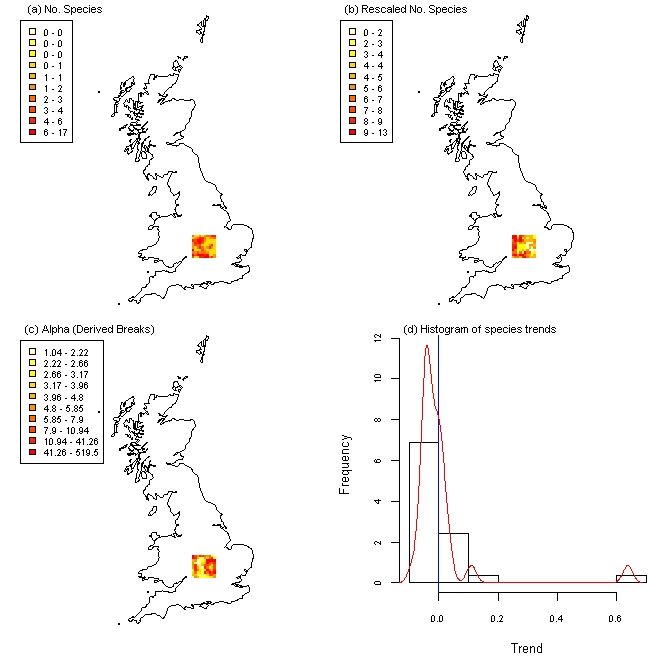
Since the example weights file we have generated in this tutorial is only for a square in the center of England you can see frescalo has limited its analysis to this area.
plot(fres_out)
Note: the plot method for frescalo objects is currently only supported for UK national grid data.
To create a distance table from a table of coordinates here is a suggested method. We will need two packages which are used in sparta, 'reshape2' and 'sp':
library(reshape2)
library(sp)
Let's create simple example dataset
location_data <- data.frame(site=c('Sparta','Athens','Delphi'),
x=c(5,6,8),
y=c(60,60,80))
print(location_data)
site x y
1 Sparta 5 60
2 Athens 6 60
3 Delphi 8 80
For the function we are going to use we need our data in a different format to this, but this is probably how you have your data. We need to change this into a matrix with two columns. The first column is the x or longitude and the second is the y or latitude.
x <- as.matrix(location_data[c('x','y')])
print(x)
x y
[1,] 5 60
[2,] 6 60
[3,] 8 80
We then have two options depending on the method we wish to use. If we wish to simply calculate the euclidean distance between sites using the same metric as is used in 'x' and 'y', we use spDists with longlat=FALSE. However, if our 'x' and 'y' represent longitude and latitude and we wish to calculate distance in kilometers using great-circle distance, we use spDists with longlat=TRUE:
# if we want distance in the same units
distances_xy<-spDists(x,y=x,longlat=FALSE)
print(distances_xy)
[,1] [,2] [,3]
[1,] 0.00000 1.00000 20.22375
[2,] 1.00000 0.00000 20.09975
[3,] 20.22375 20.09975 0.00000
# or if we are useing lat long and want distance in km
distances_latlong<-spDists(x,y=x,longlat=TRUE)
print(distances_latlong)
[,1] [,2] [,3]
[1,] 0.00000 55.79918 2232.852
[2,] 55.79918 0.00000 2231.615
[3,] 2232.85157 2231.61506 0.000
To make the distance matrix we have created readable we need to add in our site names:
row.names(distances_xy)<-location_data$site
colnames(distances_xy)<-location_data$site
print(distances_xy)
Sparta Athens Delphi
Sparta 0.00000 1.00000 20.22375
Athens 1.00000 0.00000 20.09975
Delphi 20.22375 20.09975 0.00000
This distance matrix has all the data that we need. However, the frescalo function requires the matrix to be in its long form. This is easily done using the melt() function in reshape2:
dist<-melt(distances_xy)
print(dist)
Var1 Var2 value
1 Sparta Sparta 0.00000
2 Athens Sparta 1.00000
3 Delphi Sparta 20.22375
4 Sparta Athens 1.00000
5 Athens Athens 0.00000
6 Delphi Athens 20.09975
7 Sparta Delphi 20.22375
8 Athens Delphi 20.09975
9 Delphi Delphi 0.00000
This is now in the format needed for the create_weights function and you can use your 'dist' table as described above