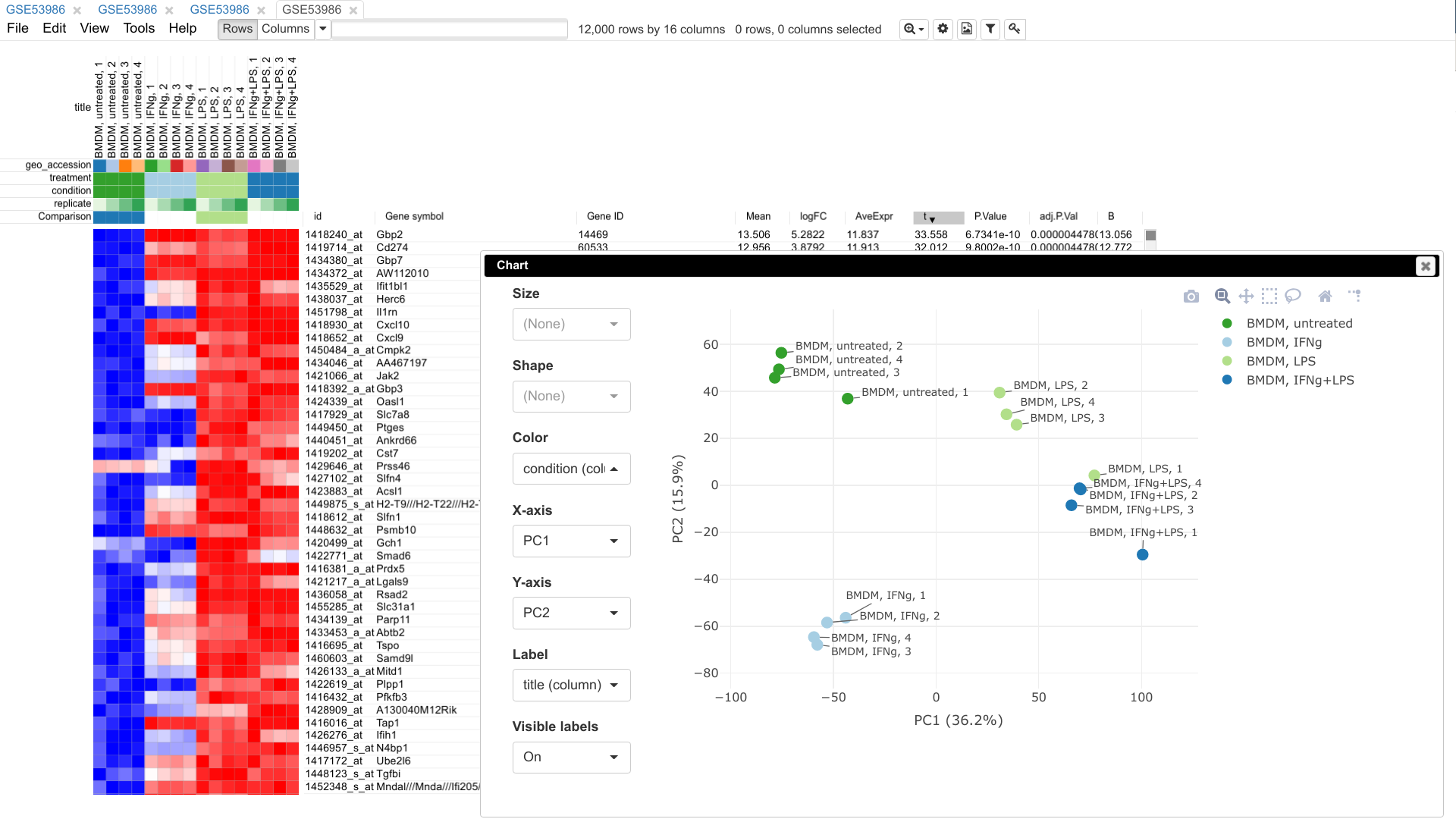
Phantasus is a web tool designed for visual and interactive gene expression analysis. In particular, it was designed to allow to go from a typical dataset to differential expression and downstream analysis in an easy and streamlined manner. For that aim, Phantasus integrates an intuitive heatmap interface with gene expression analysis tools from Bioconductor.
Main features:
- Loading public datasets from Gene Expression Omnibus with both microarrays and RNA-seq datasets (via ARCHS4) being supported.
- Differential gene expression using
limmaorDESeq2. - Publication ready plots with export to SVG: PCA plot, row profiles, box plots.
- Clustering: k-means and hierarchical.
- Gene set enrichment analysis via
fgseapackage. - Sharing session links.
Phantasus can be accessed online via its official mirror: https://alserglab.wustl.edu/phantasus.
Alternatively, phantasus can be set up locally as an R package. The latest version of Phantasus
can be installed from GitHub using devtools package.
devtools::install_github("ctlab/phantasus")There are several system packages that have to be installed on the system. The names of these packages will be displayed during installation. On Ubuntu one can install them beforehand and all together using the command:
sudo apt-get install libapparmor-dev libfontconfig1-dev libcairo2-dev libcurl4-openssl-dev pandoc libtiff5-dev libfribidi-dev libharfbuzz-dev libssl-dev libxml2-dev libprotobuf-dev protobuf-compilerFurther, the latest version of phantasus depends on rhdf5client (>= 1.25.1) from Bioconductor 3.19, which on older systems can be more convenient to install from GitHub:
devtools::install_github("vjcitn/rhdf5client")Before the first run of Phantasus you need to do an initial setup. To perform interactive setup run the following commands. We recommend to use the first option for each of the questions:
library(phantasus)
setupPhantasus()After the setup is finished, the following command will start the application at http://0.0.0.0:8000 and open it in the default browser:
servePhantasus()Please refer to the documentation for comprehensive installation and configuration instructions.
- Official mirror: https://alserglab.wustl.edu/phantasus.
- Documentation: https://ctlab.github.io/phantasus-doc.
- Bioconductor package: https://bioconductor.org/packages/phantasus.
- Docker image: https://hub.docker.com/r/alserglab/phantasus.
- Kleverov M, Zenkova D, Kamenev V, Sablina R, Artyomov M, Sergushichev A (2022). Phantasus: web-application for visual and interactive gene expression analysis. doi:10.1101/2022.12.10.519861, https://www.biorxiv.org/content/10.1101/2022.12.10.519861.