diff --git a/.github/workflows/pre-commit-checks.yml b/.github/workflows/pre-commit-checks.yml
new file mode 100644
index 0000000..2d50f95
--- /dev/null
+++ b/.github/workflows/pre-commit-checks.yml
@@ -0,0 +1,22 @@
+---
+name: tests
+
+on:
+ push:
+ branches: [main]
+ pull_request:
+ branches: [main]
+
+jobs:
+ pre_commit_checks:
+ runs-on: ubuntu-22.04
+ steps:
+ # checks out the repo
+ - uses: actions/checkout@v4
+ # setup python
+ - name: Python setup
+ uses: actions/setup-python@v5
+ with:
+ python-version: "3.11"
+ # run pre-commit
+ - uses: pre-commit/action@v3.0.1
diff --git a/.pre-commit-config.yaml b/.pre-commit-config.yaml
new file mode 100644
index 0000000..5974237
--- /dev/null
+++ b/.pre-commit-config.yaml
@@ -0,0 +1,26 @@
+# See https://pre-commit.com for more information
+# See https://pre-commit.com/hooks.html for more hooks
+default_language_version:
+ python: python3.11
+repos:
+ - repo: https://github.com/pre-commit/pre-commit-hooks
+ rev: v4.6.0
+ hooks:
+ - id: trailing-whitespace
+ - id: end-of-file-fixer
+ - id: check-added-large-files
+ - id: detect-private-key
+ - repo: https://github.com/codespell-project/codespell
+ rev: v2.3.0
+ hooks:
+ - id: codespell
+ - repo: https://github.com/executablebooks/mdformat
+ rev: 0.7.17
+ hooks:
+ - id: mdformat
+ additional_dependencies:
+ - mdformat-gfm
+ - repo: https://github.com/rhysd/actionlint
+ rev: v1.7.1
+ hooks:
+ - id: actionlint
diff --git a/CODE_OF_CONDUCT.MD b/CODE_OF_CONDUCT.MD
new file mode 100644
index 0000000..d5b0850
--- /dev/null
+++ b/CODE_OF_CONDUCT.MD
@@ -0,0 +1,131 @@
+# Contributor Covenant Code of Conduct
+
+## Our Pledge
+
+We as members, contributors, and leaders pledge to make participation in our
+community a harassment-free experience for everyone, regardless of age, body
+size, visible or invisible disability, ethnicity, sex characteristics, gender
+identity and expression, level of experience, education, socioeconomic status,
+nationality, personal appearance, race, caste, color, religion, or sexual
+identity and orientation.
+
+We pledge to act and interact in ways that contribute to an open, welcoming,
+diverse, inclusive, and healthy community.
+
+## Our Standards
+
+Examples of behavior that contributes to a positive environment for our
+community include:
+
+- Demonstrating empathy and kindness toward other people
+- Being respectful of differing opinions, viewpoints, and experiences
+- Giving and gracefully accepting constructive feedback
+- Accepting responsibility and apologizing to those affected by our mistakes,
+ and learning from the experience
+- Focusing on what is best not just for us as individuals, but for the overall
+ community
+
+Examples of unacceptable behavior include:
+
+- The use of sexualized language or imagery, and sexual attention or advances of
+ any kind
+- Trolling, insulting or derogatory comments, and personal or political attacks
+- Public or private harassment
+- Publishing others' private information, such as a physical or email address,
+ without their explicit permission
+- Other conduct which could reasonably be considered inappropriate in a
+ professional setting
+
+## Enforcement Responsibilities
+
+Community leaders are responsible for clarifying and enforcing our standards of
+acceptable behavior and will take appropriate and fair corrective action in
+response to any behavior that they deem inappropriate, threatening, offensive,
+or harmful.
+
+Community leaders have the right and responsibility to remove, edit, or reject
+comments, commits, code, wiki edits, issues, and other contributions that are
+not aligned to this Code of Conduct, and will communicate reasons for moderation
+decisions when appropriate.
+
+## Scope
+
+This Code of Conduct applies within all community spaces, and also applies when
+an individual is officially representing the community in public spaces.
+Examples of representing our community include using an official email address,
+posting via an official social media account, or acting as an appointed
+representative at an online or offline event.
+
+## Enforcement
+
+Instances of abusive, harassing, or otherwise unacceptable behavior may be
+reported to the community leaders responsible for enforcement at [gregory.way@cuanschutz.edu](mailto:gregory.way@cuanschutz.edu).
+All complaints will be reviewed and investigated promptly and fairly.
+
+All community leaders are obligated to respect the privacy and security of the
+reporter of any incident.
+
+## Enforcement Guidelines
+
+Community leaders will follow these Community Impact Guidelines in determining
+the consequences for any action they deem in violation of this Code of Conduct:
+
+### 1. Correction
+
+**Community Impact**: Use of inappropriate language or other behavior deemed
+unprofessional or unwelcome in the community.
+
+**Consequence**: A private, written warning from community leaders, providing
+clarity around the nature of the violation and an explanation of why the
+behavior was inappropriate. A public apology may be requested.
+
+### 2. Warning
+
+**Community Impact**: A violation through a single incident or series of
+actions.
+
+**Consequence**: A warning with consequences for continued behavior. No
+interaction with the people involved, including unsolicited interaction with
+those enforcing the Code of Conduct, for a specified period of time. This
+includes avoiding interactions in community spaces as well as external channels
+like social media. Violating these terms may lead to a temporary or permanent
+ban.
+
+### 3. Temporary Ban
+
+**Community Impact**: A serious violation of community standards, including
+sustained inappropriate behavior.
+
+**Consequence**: A temporary ban from any sort of interaction or public
+communication with the community for a specified period of time. No public or
+private interaction with the people involved, including unsolicited interaction
+with those enforcing the Code of Conduct, is allowed during this period.
+Violating these terms may lead to a permanent ban.
+
+### 4. Permanent Ban
+
+**Community Impact**: Demonstrating a pattern of violation of community
+standards, including sustained inappropriate behavior, harassment of an
+individual, or aggression toward or disparagement of classes of individuals.
+
+**Consequence**: A permanent ban from any sort of public interaction within the
+community.
+
+## Attribution
+
+This Code of Conduct is adapted from the [Contributor Covenant][homepage],
+version 2.1, available at
+[https://www.contributor-covenant.org/version/2/1/code_of_conduct.html][v2.1].
+
+Community Impact Guidelines were inspired by
+[Mozilla's code of conduct enforcement ladder][mozilla coc].
+
+For answers to common questions about this code of conduct, see the FAQ at
+[https://www.contributor-covenant.org/faq][faq]. Translations are available at
+[https://www.contributor-covenant.org/translations][translations].
+
+[faq]: https://www.contributor-covenant.org/faq
+[homepage]: https://www.contributor-covenant.org
+[mozilla coc]: https://github.com/mozilla/diversity
+[translations]: https://www.contributor-covenant.org/translations
+[v2.1]: https://www.contributor-covenant.org/version/2/1/code_of_conduct.html
diff --git a/README.md b/README.md
index 81c8cad..779e4a3 100644
--- a/README.md
+++ b/README.md
@@ -1,17 +1,19 @@
-# Playbooks
+# Lab Playbooks
Playbooks relevant to Way Lab operations.
+See the GitHub Pages version of this content here: [https://wayscience.github.io/playbooks/](https://wayscience.github.io/playbooks/).
+
Top-level playbooks have information which may be relevant to other computational labs.
Playbooks in the `/internal` folder have information specific to the Way Lab.
-| Playbook | Type | Description |
-|-------------------------|-----------------------|------------------------------------|
-| [Code Strategy](code_strategy.md) | General | Strategy for using and writing code in a scientific setting. |
-| [Conference Playbook](conference_playbook.md) | General | Playbook for selecting and participating in conferences. |
-| [Data Strategy](data_strategy.md) | General | Strategy for using and storing data; includes our 3 data perspectives. |
-| [GitHub Strategy](github_strategy.md) | General | Strategy for using GitHub efficiently; details our 3 types of repositories. |
-| [Calendar Playbook](internal/calendar_playbook.md) | Internal | Playbook for joining and using Way Lab calendar. |
-| [Computer Setup](internal/computer_setup.md) | Internal | Instructions for lab computer setup. |
-| [Onboarding](internal/onboarding.md) | Internal | Onboarding materials for the Way Lab. |
-| [Remote Access](internal/remote_access.md) | Internal | Instructions for remotely connecting to and using Way Lab computers. |
+| Playbook | Type | Description |
+| -------------------------------------------------- | -------- | --------------------------------------------------------------------------- |
+| [Code Strategy](code_strategy.md) | General | Strategy for using and writing code in a scientific setting. |
+| [Conference Playbook](conference_playbook.md) | General | Playbook for selecting and participating in conferences. |
+| [Data Strategy](data_strategy.md) | General | Strategy for using and storing data; includes our 3 data perspectives. |
+| [GitHub Strategy](github_strategy.md) | General | Strategy for using GitHub efficiently; details our 3 types of repositories. |
+| [Calendar Playbook](internal/calendar_playbook.md) | Internal | Playbook for joining and using Way Lab calendar. |
+| [Computer Setup](internal/computer_setup.md) | Internal | Instructions for lab computer setup. |
+| [Onboarding](internal/onboarding.md) | Internal | Onboarding materials for the Way Lab. |
+| [Remote Access](internal/remote_access.md) | Internal | Instructions for remotely connecting to and using Way Lab computers. |
diff --git a/code_strategy.md b/code_strategy.md
index d39ee38..6b13f9b 100644
--- a/code_strategy.md
+++ b/code_strategy.md
@@ -1,5 +1,7 @@
+# Code Strategy
+
We are biomedical data scientists who use code to answer biological hypotheses and explore biological data.
-It is therefore important to define how we use, write, and share code.
+It is therefore important to define how we use, write, and share code.
## 1. Using code
@@ -38,7 +40,7 @@ Good code is readable.
Based on the logic, variable names, and documentation, anyone (especially you!) should be able to understand what your code is doing.
Readability also means standard formatting.
We always lint our code, and use automatic formatting wherever possible (e.g. [python black](https://github.com/psf/black)).
-We use pre-commit hooks to ensure our code follows conventions before commiting.
+We use pre-commit hooks to ensure our code follows conventions before committing.
### 3. Code should be well organized (maintainable)
@@ -54,12 +56,10 @@ Waylab code should (usually) follow these conventions for readable, functional c
Note that these conventions will change as the field of biomedical data science evolves and improves.
1. Use [formatted string literals (f-strings)](https://docs.python.org/3/tutorial/inputoutput.html#tut-f-strings) to concatenate or insert information into strings.
+1. Use [pathlib](https://docs.python.org/3/library/pathlib.html) to represent file-system paths.
+1. Save intermediate files in parquet (`.parquet`) format when possible.
-2. Use [pathlib](https://docs.python.org/3/library/pathlib.html) to represent file-system paths.
-
-3. Save intermediate files in parquet (`.parquet`) format when possible.
-
-In general, we follow these [10 simple rules for reproducible computational research](https://doi.org/10.1371/journal.pcbi.1003285).
+In general, we follow these [10 simple rules for reproducible computational research](https://doi.org/10.1371/journal.pcbi.1003285).
## 3. Sharing code
@@ -95,7 +95,6 @@ Reviewing code can be tricky.
It is important to understand someone else's code and goals, but also to let their own style shine through their work.
We strive for individuality, understanding, and accuracy, so if you are able to spot a bug (or the potential for one), please highlight in a concise review.
-
### Code review checklist
- [ ] Do you understand the code’s intention?
@@ -121,7 +120,7 @@ We strive for individuality, understanding, and accuracy, so if you are able to
Having someone else look at your code can be scary and intimidating!
The Waylab PI (Greg) will review your first couple pull requests to ease you into this process. However, with the lab growing and projects expanding, lab-wide pull requests are necessary.
Work with Greg to find a pull request buddy, who will quickly become an expert on, and a cheerleader for, your project.
-Alternatively, post your pull request in the `pull-request-review` Discord channel.
+Alternatively, post your pull request in the `pull-request-review` Discord channel.
Please make your pull requests easy to review.
They should be concise, clearly documented, and only change a minimum number of topics (ideally, 1 core change per PR).
@@ -149,14 +148,14 @@ The methodology is as follows:
1. Create a new branch from _main_ for the entire refactor
-2. Create a new branch from the entire refactor branch for a specific change
+1. Create a new branch from the entire refactor branch for a specific change
-3. File a PR from this change-specific branch into the entire refactor branch
+1. File a PR from this change-specific branch into the entire refactor branch
-4. Someone will review your PR and after approval…
+1. Someone will review your PR and after approval…
-5. Squash merge the change-specific branch into the entire refactor branch
+1. Squash merge the change-specific branch into the entire refactor branch
-6. Repeat 2-5 for all “chunks of change”
+1. Repeat 2-5 for all “chunks of change”
-7. Finally, once all the changes are introduced, file a PR from the entire refactor branch into _main_!
+1. Finally, once all the changes are introduced, file a PR from the entire refactor branch into _main_!
diff --git a/conference_playbook.md b/conference_playbook.md
index b46dbc3..e61530c 100644
--- a/conference_playbook.md
+++ b/conference_playbook.md
@@ -1,30 +1,23 @@
+# Conference Strategy
+
## Purpose
Scientific conferences are an important part of the scientific enterprise.
We attend scientific conferences to disseminate our work, exchange ideas, learn about new fields and approaches, meet new people, catch up with old friends, hone our scientific communication, and have fun.
-
## How to decide attendance
So you’re thinking of attending a conference? Great! Ask yourself these questions to see if it makes sense to attend:
1. Is the conference reputable and generally well-attended?
-
-2. Are you interested in the topic and program?
-
-3. Is the topic directly relevant, directly adjacent, or complementary to your work?
-
-4. Would attendance advance your work and/or career in a meaningful way?
-
-5. Are there opportunities for you to present your work?
-
-6. Are there opportunities for you to receive valuable feedback on your work?
-
-7. Is now an appropriate time to talk about your work?
-
-8. Would attending the conference significantly disrupt any other existing commitments/responsibilities?
-
-9. Are there remote attendance options?
+1. Are you interested in the topic and program?
+1. Is the topic directly relevant, directly adjacent, or complementary to your work?
+1. Would attendance advance your work and/or career in a meaningful way?
+1. Are there opportunities for you to present your work?
+1. Are there opportunities for you to receive valuable feedback on your work?
+1. Is now an appropriate time to talk about your work?
+1. Would attending the conference significantly disrupt any other existing commitments/responsibilities?
+1. Are there remote attendance options?
It is not always an appropriate time to attend a conference.
Sometimes, our time is more valuable wrapping up a project, focusing on new work, or preparing for an even greater opportunity.
@@ -37,117 +30,76 @@ In nearly every case, it only makes sense to attend a conference if you are pres
In order of value, presentations include:
1. Invited keynote
-
-2. Invited seminar
-
-3. Organizing committee for workshop/panel
-
-4. Accepted seminar
-
-5. Lightning talk
-
-6. Poster
+1. Invited seminar
+1. Organizing committee for workshop/panel
+1. Accepted seminar
+1. Lightning talk
+1. Poster
The invited talks sometimes may include an honorarium.
-
## Conferences on our radar
### CytoData
-
-
- This is our field’s primary conference; the work is directly relevant to the lab, many of these folks are primary users of our software.
-
- A small, close-knit community that is rapidly expanding.
-
- The conference is international; one year it is in the US, the next year it is in Europe.
-
- There are not always poster sessions, the format changes from year-to-year.
-
### SBI2
-
-
- This is a directly adjacent field that is often more focused on image data collection than image data analysis (although recently the analysis has come into greater focus).
-
- The attendees of this conference are mixed evenly between academia and industry.
-
- We would love for this community to use our software.
-
- This conference is always in Boston.
-
### SLAS
-
-
- This is a bit further away from our field than SBI2, and contains more industry attendees.
-
- The topics are very diverse, but focused on biological imaging.
-
- We would love for this community to use our software.
-
### Pacific Symposium on Biocomputing
-
-
- A general-purpose bioinformatics and computational biology conference that is typically very well attended by senior academics and influential thinkers.
-
- The conference is always on the Big Island of Hawaii, occurring yearly in January.
-
- The topics are always topical, and change yearly.
-
- It typically only makes sense to attend this conference if you have an accepted paper, accepted talk, or organize a workshop.
-
- Travel awards are available, and presenting students have great odds at acceptance.
-
### NF Conference
-
-
- This is a biological domain conference focused on Neurofibromatosis (NF)
-
- We attend domain conferences like this one when we have new and unique insights and/or innovative approaches to share with the community
-
### Neurips
-
-
- This is a huge, general machine learning conference
-
- Often there are biomedical application workshops, which we can present our work in
-
- Many sessions are helpful and can directly inspire future projects
-
- Travel awards are available, so please apply
-
### CVPR
-
-
- One of the top computer vision conferences in the world
-
- Very similar to Neurips, but focused on computer vision theory, models, and applications
-
- There are general paper submissions available, but this is very competitive
### ASCB
- American Society for Cell Biology
-
- Great conference that focuses on cell biology processes and mechanisms
-
- There is a strong emphasis on microscopy techniques and applications as this is a powerful tool for studying cells
-
- Larger conference
-
## How to guide (attendance and reimbursement)
> :exclamation: **Waylab Specific**: This may not apply to other labs!
@@ -155,6 +107,7 @@ The invited talks sometimes may include an honorarium.
The lab will cover all reasonable costs for conference attendance.
This includes transportation, meals, registration, and incidentals.
Please ask questions.
+
**_Please save receipts as they could be required._**
We use a service called [Concur](https://www.ucdenver.edu/offices/provost/office-of-the-provost/hr-and-finance-toolbox-home/procurement-and-travel/travel#ac-pre-travel-0) to track reimbursements and travel requests.
@@ -165,70 +118,36 @@ Concur is where you will document all conference related expenses that require r
The steps for paying/reimbursement include:
1. Decide if attendance makes sense and is authorized
-
-2. File a “Travel request” on Concur
-
- 1. Follow this step-by-step guide here:
-
- 2. See for details
-
-3. Check if there is a travel award, and apply if so!
-
+1. File a “Travel request” on Concur
+ 1. Follow this step-by-step guide here:
+ 1. See for details
+1. Check if there is a travel award, and apply if so!
1. This is important! It looks great on your CV and saves the lab money.
-
-4. Register for the conference
-
+1. Register for the conference
1. **Note:** Sometimes the order of this step is slightly different, but this is recommended to do after getting your pre-travel request approved.
-
- 2. Sometimes registration requires a first step of professional organization membership
-
- 3. There are two methods for registering for conferences:
-
+ 1. Sometimes registration requires a first step of professional organization membership
+ 1. There are two methods for registering for conferences:
1. Book yourself, save receipts
-
1. Reimbursed later, after travel
-
- 2. Book with Jenna Tomkinson using the lab procurement card
-
+ 1. Book with Jenna Tomkinson using the lab procurement card
1. Paid directly with lab funds
-
- 2. Make sure there is **NO TAX**
-
-5. Book airfare (if applicable). Please be reasonable in terms of cost!
-
+ 1. Make sure there is **NO TAX**
+1. Book airfare (if applicable). Please be reasonable in terms of cost!
1. Use the Concur “Travel” tab and use the “Mixed Flight/Train Search” on the left hand side to look up flights.
-
- 2. Once you have add the information for departure and arrival dates, it will provide all flights available from all airlines
-
+ 1. Once you have add the information for departure and arrival dates, it will provide all flights available from all airlines
1. **Note:** It will show you which airlines that the university prefers and breaks down the available flights by how many stops there are.
-
- 3. Once you find the flights you want, follow ALL STEPS to the end or the reservation will be canceled.
-
- 4. The lab typically does not cover rental cars
-
-6. Book lodging (if applicable)
-
- 1. Often the conference offers a discounted hotel block/code.
- Use this code and book early to ensure accommodations
-
- 2. You will need to book this yourself and pay this out of pocket
-
+ 1. Once you find the flights you want, follow ALL STEPS to the end or the reservation will be canceled.
+ 1. The lab typically does not cover rental cars
+1. Book lodging (if applicable)
+ 1. Often the conference offers a discounted hotel block/code. Use this code and book early to ensure accommodations
+ 1. You will need to book this yourself and pay this out of pocket
1. Reimbursed later after travel
-
-7. Save receipts, save receipts, save receipts!
-
+1. Save receipts, save receipts, save receipts!
1. File for reimbursement on concur immediately upon return.
-
-8. **AFTER TRAVEL** (e.g., once you come back), you will go back into Concur to your approved request to add all expenses.
-
+1. **AFTER TRAVEL** (e.g., once you come back), you will go back into Concur to your approved request to add all expenses.
1. Typical expenses include:
-
1. Per-diem (for food)
-
- 2. Hotel/Lodging
-
- 3. Registration (if paid out-of-pocket)
-
- 4. Transportation (if paid out-of-pocket)
-
- 5. Poster printing
+ 1. Hotel/Lodging
+ 1. Registration (if paid out-of-pocket)
+ 1. Transportation (if paid out-of-pocket)
+ 1. Poster printing
diff --git a/data_strategy.md b/data_strategy.md
index 79c2a7d..5d88233 100644
--- a/data_strategy.md
+++ b/data_strategy.md
@@ -1,5 +1,7 @@
+# Data Strategy
+
A suitable and flexible data management plan is essential for effective and trustworthy science.
-Our goals with this strategy is to maximize access, understanding, analysis speed, and provenance while reducing access barriers, unnecessary storage bloat, and cost.
+Our goals with this strategy is to maximize access, understanding, analysis speed, and provenance while reducing access barriers, unnecessary storage bloat, and cost.
## 1. Data perspectives
@@ -7,14 +9,13 @@ We think of data from three different perspectives:
1. Level
-2. Origin
+1. Origin
-3. Flow
+1. Flow
Each perspective requires different considerations for storage, access, and provenance management.
Management practices for microscopy images are related to other data types, with some nuance.
-
### Level
Your data level indicates the processing stage.
@@ -25,17 +26,15 @@ With biological data, there are many different kinds of intermediate data.
Intermediate data are typically different sizes and thus have different storage requirements.
Each intermediate data type requires unique considerations for access frequency, dissemination, and versioning.
-
### Origin
Where your data come from also requires unique management policies.
We use data originating from collaborators (both academic and industry) and data already in the public domain.
-Eventually, we will use data that we ourselves collect, but for the moment, we can ignore this origin category.
+Eventually, we will use data that we ourselves collect, but for the moment, we can ignore this origin category.
We need to consider access requirements and restrictions, particularly when using collaborator data.
When storing restricted data, it is helpful to remember that all data will eventually be in the public domain.
-
### Flow
Besides the most raw form, data are dynamic and pluripotent; always awaiting new and improved processing capabilities.
@@ -56,35 +55,34 @@ We consider three categories of potential storage solutions for data:
1. Internal hard drive
- 2. External hard drive
+ 1. External hard drive
-2. Cloud storage
+1. Cloud storage
1. Image Data Resource (IDR)
- 2. Amazon/GC/Azure
+ 1. Amazon/GC/Azure
- 3. Figshare/Figshare+
+ 1. Figshare/Figshare+
- 4. Zenodo
+ 1. Zenodo
- 5. Github
+ 1. Github
- 6. Github LFS
+ 1. Github LFS
- 7. DVC
+ 1. DVC
- 8. Colorado local cluster
+ 1. Colorado local cluster
- 9. One Drive/Dropbox/Google drive
+ 1. One Drive/Dropbox/Google drive
-3. No storage
+1. No storage
1. Immediate deletion
Each storage solution has trade-offs in terms of longevity, access, usage speed, version control, size restrictions, and cost (**Table 1**).
-
## 3. Microscopy Data Levels
From the raw microscopy image to the variable intermediate data types including single cell and bulk embeddings, each data level has unique data storage considerations (**Figure 1**).
@@ -98,23 +96,23 @@ We apply representation learning pipelines to extract morphology features from s
Finally, we apply an aggregated bulk pipeline to turn the single cell morphology embeddings into aggregated bulk embeddings.
Importantly, we have different short, mid, and long term storage and sharing solutions for each data type.
-| | | | | | | |
-| -------------------------------------------------------------------------------------- | ------------- | ------------------- | --------------- | --------------- | -------------------------------------------------------------------------------------------------------------------------------------------- | -------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
-| **Solution** | **Longevity** | **Version Control** | **Access** | **Usage speed** | **Size limits** | **Cost** |
-| [Internal hard drive](https://www.dpbestflow.org/data-storage-hardware/hard-drive-101) | Intermediate | No | Private | Instant | <= 18TB (Total) | \~$15 per TB one time cost ([Details](https://diskprices.com/)) |
-| External hard drive | High | No | Private | Download | <= 18TB (Total) | \~$15 per TB one time cost ([Details](https://diskprices.com/)) |
-| [IDR](https://idr.openmicroscopy.org/) | High | Yes | Public | Download | >= 2TB (Per dataset) | Free |
-| AWS/GC/Azure | Low | Yes | Public/Private | Instant | >= 2TB (Per dataset) | $0.02 - $0.04 per GB / Month ($40 to $80 per month per 2TB dataset) |
-| [Figshare](https://figshare.com/) | High | Yes | Public | Download | 20GB (Total) | Free ([Details](https://help.figshare.com/article/figshare-account-limits)) |
-| [Figshare+](https://knowledge.figshare.com/plus) | High | Yes | Public | Download | 250GB > x > 5TB (Per dataset) | $745 > x > $11,860 one time cost ([Details](https://knowledge.figshare.com/plus)) |
-| [Zenodo](https://zenodo.org/) | High | Yes | Public | Download | >= 50GB (Per dataset) | Free ([Details](https://help.zenodo.org/)) |
-| Github | High | Yes | Public/Private | Instant | >= 100MB (Per file) ([Details](https://docs.github.com/en/repositories/working-with-files/managing-large-files/about-large-files-on-github)) | Free |
-| Github LFS | Intermediate | Yes | Public/Private | Instant | >= 2GB (up to 5GB for paid plans) | 50GB data pack for $5 per month ([Details](https://docs.github.com/en/billing/managing-billing-for-git-large-file-storage/about-billing-for-git-large-file-storage)) |
-| DVC | Intermediate | Yes | Public/Private | Download | None | Cost of linked service (AWS/Azure/GC) |
-| One drive | Low | Yes | Private | Instant | >= 5TB (Total) | Free to AMC |
-| Dropbox | Low | Yes | Public/Private | Instant | >= 5TB (Total) | $12.50 per user / month ([Details](https://www.dropbox.com/plans)) |
-| Google drive | Low | Yes | Public/Private | Instant | >= 5TB (Total) | $25 per month ([Details](https://one.google.com/about/plans)) |
-| Local cluster (RMACC) | Intermediate | No | Private | Instant | | |
-| Immediate deletion | None | None | None | None | None | None |
+| | | | | | | |
+| -------------------------------------------------------------------------------------- | ------------- | ------------------- | -------------- | --------------- | -------------------------------------------------------------------------------------------------------------------------------------------- | -------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
+| **Solution** | **Longevity** | **Version Control** | **Access** | **Usage speed** | **Size limits** | **Cost** |
+| [Internal hard drive](https://www.dpbestflow.org/data-storage-hardware/hard-drive-101) | Intermediate | No | Private | Instant | \<= 18TB (Total) | ~$15 per TB one time cost ([Details](https://diskprices.com/)) |
+| External hard drive | High | No | Private | Download | \<= 18TB (Total) | ~$15 per TB one time cost ([Details](https://diskprices.com/)) |
+| [IDR](https://idr.openmicroscopy.org/) | High | Yes | Public | Download | >= 2TB (Per dataset) | Free |
+| AWS/GC/Azure | Low | Yes | Public/Private | Instant | >= 2TB (Per dataset) | $0.02 - $0.04 per GB / Month ($40 to $80 per month per 2TB dataset) |
+| [Figshare](https://figshare.com/) | High | Yes | Public | Download | 20GB (Total) | Free ([Details](https://help.figshare.com/article/figshare-account-limits)) |
+| [Figshare+](https://knowledge.figshare.com/plus) | High | Yes | Public | Download | 250GB > x > 5TB (Per dataset) | $745 > x > $11,860 one time cost ([Details](https://knowledge.figshare.com/plus)) |
+| [Zenodo](https://zenodo.org/) | High | Yes | Public | Download | >= 50GB (Per dataset) | Free ([Details](https://help.zenodo.org/)) |
+| Github | High | Yes | Public/Private | Instant | >= 100MB (Per file) ([Details](https://docs.github.com/en/repositories/working-with-files/managing-large-files/about-large-files-on-github)) | Free |
+| Github LFS | Intermediate | Yes | Public/Private | Instant | >= 2GB (up to 5GB for paid plans) | 50GB data pack for $5 per month ([Details](https://docs.github.com/en/billing/managing-billing-for-git-large-file-storage/about-billing-for-git-large-file-storage)) |
+| DVC | Intermediate | Yes | Public/Private | Download | None | Cost of linked service (AWS/Azure/GC) |
+| One drive | Low | Yes | Private | Instant | >= 5TB (Total) | Free to AMC |
+| Dropbox | Low | Yes | Public/Private | Instant | >= 5TB (Total) | $12.50 per user / month ([Details](https://www.dropbox.com/plans)) |
+| Google drive | Low | Yes | Public/Private | Instant | >= 5TB (Total) | $25 per month ([Details](https://one.google.com/about/plans)) |
+| Local cluster (RMACC) | Intermediate | No | Private | Instant | | |
+| Immediate deletion | None | None | None | None | None | None |
**Table 1**: Tradeoffs and considerations for data storage solutions.
diff --git a/github_strategy.md b/github_strategy.md
index 949d99e..d42039d 100644
--- a/github_strategy.md
+++ b/github_strategy.md
@@ -1,3 +1,5 @@
+# GitHub Strategy
+
Sharing resources is fundamental to advancing biomedical research.
We share code, software, analyses, results, and data (or data access instructions) using Github.
Our Github page is here:
@@ -8,9 +10,9 @@ There are three categories of github repositories, each with specific design pat
1. Software
-2. Analysis
+1. Analysis
-3. Data
+1. Data
We will outline each of the three below.
@@ -44,14 +46,14 @@ Examples: [pycytominer](https://github.com/cytomining/pycytominer), [CytoSnake](
As biomedical data scientists, analysis repositories are the most common type of repository for our group.
When most people think about a github repository, they are likely not thinking about storing a reproducible analysis.
-Nevertheless, this approach is gaining in popularity.
+Nevertheless, this approach is gaining in popularity.
An analysis repository contains a collection of analyses linked together by the name of the project, which should be descriptive of the specific set of interrelated hypotheses.
Analysis repositories contain numbered folders, or modules, that have a sequential relationship. For example, the first step in most analysis repositories is data access.
Therefore, the first module is often called \`0.download-data\`, and it contains instructions on how to access the data you use in your analysis.
The subsequent modules all use these data in various ways; often exploring the data or testing various hypotheses.
-We also include code for generating all publication quality figures as the last module.
+We also include code for generating all publication quality figures as the last module.
We use software libraries in these analyses, and we save specific versions in conda environment files or docker containers.
We also must specify the license, present a clear README with reproducibility/usage instructions, a brief summary of the results, and citation instructions.
@@ -59,9 +61,9 @@ We also must specify the license, present a clear README with reproducibility/us
Analysis repositories must be included in our publications, and we consider them to be the ground truth methods section. The analyses must be fully reproducible (we aim for [Gold Reproducibility](https://www.nature.com/articles/s41592-021-01256-7#Sec4)) and open source.
For each submission and subsequent resubmission, we generate specific “github releases”.
We generate direct object identifiers (DOI) for these releases using Zenodo ([instructions](https://docs.github.com/en/repositories/archiving-a-github-repository/referencing-and-citing-content)).
-We do not restrict ourselves to the one-repository per-paper strategy, instead, we opt for the one-release per-submission strategy (which can span multiple publications).
+We do not restrict ourselves to the one-repository per-paper strategy, instead, we opt for the one-release per-submission strategy (which can span multiple publications).
-Examples: [phenotypic\_profiling\_model](https://github.com/WayScience/phenotypic_profiling_model), [gene\_dependency\_representations](https://github.com/WayScience/gene_dependency_representations)
+Examples: [phenotypic_profiling_model](https://github.com/WayScience/phenotypic_profiling_model), [gene_dependency_representations](https://github.com/WayScience/gene_dependency_representations)
### 3. Data repository
@@ -76,7 +78,8 @@ We also make sure that this link between the pipeline and data is itself version
This process ensures that our data can be easily reprocessed by updated bioinformatics methods with new and improved methods.
-Examples from NF1 publication:
+Examples from NF1 publication:
+
- ML and bioinformatics pipeline: [nf1_schwann_cell_morphology_signature](https://github.com/WayScience/nf1_schwann_cell_morphology_signature)
- Data and image profiling: [nf1_schwann_cell_painting_data](https://github.com/WayScience/nf1_schwann_cell_painting_data)
@@ -86,24 +89,22 @@ There are important considerations to make when creating, contributing, and shar
## 3. Github repositories as lab notebooks
-We use GitHub repositories functionally as electronic lab notebooks.
+We use GitHub repositories functionally as electronic lab notebooks.
## 4. Pull request peer review
-The contributor can only merge code into WayScience (or affiliate \[e.g., Cytomining]) repositories after it is approved by at least one Way lab arborist. ✅
+The contributor can only merge code into WayScience (or affiliate \[e.g., Cytomining\]) repositories after it is approved by at least one Way lab arborist. ✅
This process has many direct and indirect benefits.
-
### Direct benefits of pull request review
- Only high-quality, understandable code exists in the WayScience GitHub organization.
-- We minimize future pain involved with perpetuating bugs over time (i.e., technical debt) which will lead to inaccurate or misleading interpretations.
+- We minimize future pain involved with perpetuating bugs over time (i.e., technical debt) which will lead to inaccurate or misleading interpretations.
- While pull request review slows progress in the short term, the added benefits of sustainability more than make up for this time lost in the long term.
-
### Indirect benefits of pull request review
#### To the scientist writing the code (the writer)
@@ -112,7 +113,6 @@ This process has many direct and indirect benefits.
- Encourages committing early and often, which reduces the chance of unintentional code deletion
-
#### To the scientist reviewing the code (the reviewer)
- Adds accountability, which also encourages higher quality work.
@@ -121,7 +121,6 @@ This process has many direct and indirect benefits.
- Remaining up to date on the progress of the project, which increases collaboration and comradery in the Lab.
-
### Step-by-step process
1. The writer files a **small, bite-sized** pull request 🪥
@@ -130,23 +129,23 @@ This process has many direct and indirect benefits.
- The smaller the better; it is WAY easier to review if small
-2. The writer adds details to the pull request in markdown format
+1. The writer adds details to the pull request in markdown format
- Justifies the rationale for why the pull request exists
- - \[Optional] Encourages the reviewer to pay particular attention to any aspect
+ - \[Optional\] Encourages the reviewer to pay particular attention to any aspect
- If the pull request contains a figure, add the figure
-3. The writer posts a link to the pull request to the #pull-request-review discord channel
+1. The writer posts a link to the pull request to the #pull-request-review discord channel
- Often, the writer knows who should take a look. Tag this person.
-4. The reviewer will see the post on the discord channel and determine if they can (and are suitable) to review
+1. The reviewer will see the post on the discord channel and determine if they can (and are suitable) to review
- The reviewer can post to the discord channel claiming the review
-5. The reviewer will review the pull request
+1. The reviewer will review the pull request
- Review with the desire:
@@ -159,33 +158,51 @@ This process has many direct and indirect benefits.
- Review to pass along tips along any of the above axes
- **Note:** Standards change over time, and we set our own standards internally.
- We do not prescribe a standard beyond the three items above.
+ We do not prescribe a standard beyond the three items above.
-6. The reviewer will submit their review
+1. The reviewer will submit their review
- This review may, or may not, include an approval ✅
-7. Regardless of approval, the writer will respond to the reviewers comments (if there are any), and a discussion may ensue.
+1. Regardless of approval, the writer will respond to the reviewers comments (if there are any), and a discussion may ensue.
- **Note**: Most often, the writer is the project owner.
- This means they have final say in what changes are incorporated.
+ This means they have final say in what changes are incorporated.
- In rare cases, if there is a disagreement that cannot be resolved, involve the PI.
-8. After approval, the writer will squash+merge the pull request into main
+1. After approval, the writer will squash+merge the pull request into main
-| | | Software | Analysis | Data |
+| | | Software | Analysis | Data |
| ---------------- | ---------------------------------- | :-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------: | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | ---------------------------------------------------------------------------------------------------------------- |
-| Creating | _Name_ | Up to the primary developer, PI has veto power | Up to the primary developer, PI has veto power | Descriptive of the project, typically the project is named well before the data are collected |
-| | _Location_ | Most often lab org | Lab org | Lab org or pre-defined by project scope |
-| | _Initial files_ | README.md, LICENSE.md, CONTRIBUTING.md, CODE\_OF\_CONDUCT.md | | |
+| Creating | _Name_ | Up to the primary developer, PI has veto power | Up to the primary developer, PI has veto power | Descriptive of the project, typically the project is named well before the data are collected |
+| | _Location_ | Most often lab org | Lab org | Lab org or pre-defined by project scope |
+| | _Initial files_ | README.md, LICENSE.md, CONTRIBUTING.md, CODE_OF_CONDUCT.md | | |
| **Contributing** | _Location_ | Fork of matching lab repo, in specific feature-named branches | | |
-| | _Tests_ | Maximal code coverage and continuous integration with Github Actions | Encouraged for critical analysis code used over and over again | Not required; software tested elsewhere, all pipeline steps reviewed and intermediate data tested in other repos |
+| | _Tests_ | Maximal code coverage and continuous integration with Github Actions | Encouraged for critical analysis code used over and over again | Not required; software tested elsewhere, all pipeline steps reviewed and intermediate data tested in other repos |
| | _Code review\*_ | Limit changes per pull request (e.g. one core change); at least one approval required by project maintainer prior to pull request merge or as described in repo contributing guidelines | One analysis (or less) per pull request; one approval by lab member (or collaborator) required | One approval by a lab member (or collaborator) required. Try to keep contributions per pull request small |
| | _Adding files_ | Everything from feature-name branches; Limit large files and do not include them in distributions | Everything from feature-name branches; Depending on size, see [Data Strategy](https://docs.google.com/document/d/1qRz-hCTvZIZIDP4a7wLtQfbmOhKyGA51_ncKVzAnAHU/edit?usp=sharing) | |
-| | _Adding dependencies_ | Limit dependencies, add specific version(s) to requirements file | Limit dependencies, add specific version to environment file | Only pipeline and standard data visualization/exploration software |
+| | _Adding dependencies_ | Limit dependencies, add specific version(s) to requirements file | Limit dependencies, add specific version to environment file | Only pipeline and standard data visualization/exploration software |
| | _Organization_ | Standard package organization | Module-based sequences that depend on prior steps | Store critical intermediate data in different forms, or, if too large, include access instructions |
-| **Sharing** | _Version control and citable code_ | Maintain version on common package installers (e.g. pip, conda); include citation instructions | Mint a DOI using zenodo per release; Github releases track paper submissions | Mint a DOI using zenodo per release; Github releases track full pipelines |
+| **Sharing** | _Version control and citable code_ | Maintain version on common package installers (e.g. pip, conda); include citation instructions | Mint a DOI using zenodo per release; Github releases track paper submissions | Mint a DOI using zenodo per release; Github releases track full pipelines |
**Table 1:** Creating, contributing, and sharing software, analysis, and data repositories.
-*See details on code review below
+\*See details on code review below
+
+## Using GitHub as a Web Host (GitHub Pages)
+
+GitHub provides the ability to host web pages through a feature called [GitHub Pages](https://pages.github.com/).
+Websites built with GitHub Pages are generally considered ["static sites"](https://en.wikipedia.org/wiki/Static_web_page).
+By default, GitHub Pages uses the [Jekyll](https://jekyllrb.com/) static site generator.
+You may also configure other static sites by using the [`.nojekyll` file](https://github.blog/news-insights/bypassing-jekyll-on-github-pages/) at the root of your GitHub Pages content.
+
+### Configuring GitHub Pages with Defaults
+
+1. Go to your repository on GitHub.
+1. Ensure there are markdown (`.md` files) within the root of the repository (for example, a `readme.md` file).
+1. Click on the "Settings" tab (located near the top of the repository).
+1. Scroll down to the "Pages" section.
+1. Under "Branch," select `main` (or your default branch) from the dropdown, and leave the folder as `/root`.
+1. Click "Save" and GitHub Pages will be enabled using the defaults (note: it takes a few moments for the initial deployment and URL to appear after this).
+
+Your site will be live at `https://.github.io//`.
diff --git a/internal/calendar_playbook.md b/internal/calendar_playbook.md
index b5edd42..f21ad15 100644
--- a/internal/calendar_playbook.md
+++ b/internal/calendar_playbook.md
@@ -1,19 +1,20 @@
+# Calendar Playbook
+
Having a shared calendar for a lab can be a game changer in organization.
-By having every individual in the lab add their events into one calendar, then spontaneous drop-ins and coordinating schedules is a breeze!
+By having every individual in the lab add their events into one calendar, then spontaneous drop-ins and coordinating schedules is a breeze!
## 1) Get added to the Way Lab Group
Firstly, everyone should be a part of the **“Way Lab”** Outlook group.
You can check this by going to the **“People”** tab in Outlook (far left side) and clicking **“Member”** under the **“Group”** section to view all of the groups you are a member of.
-If you do not see the Way Lab group, then please contact Jenna Tomkinson (**bbyy\_j22**) on Discord so she can add you.
+If you do not see the Way Lab group, then please contact Jenna Tomkinson (**bbyy_j22**) on Discord so she can add you.
## 2) View the calendar on Outlook
Once you are a member, you can locate the Way Lab calendar by scrolling down on the left side where **“My calendars”** directory is located to the **“Groups”** directory.
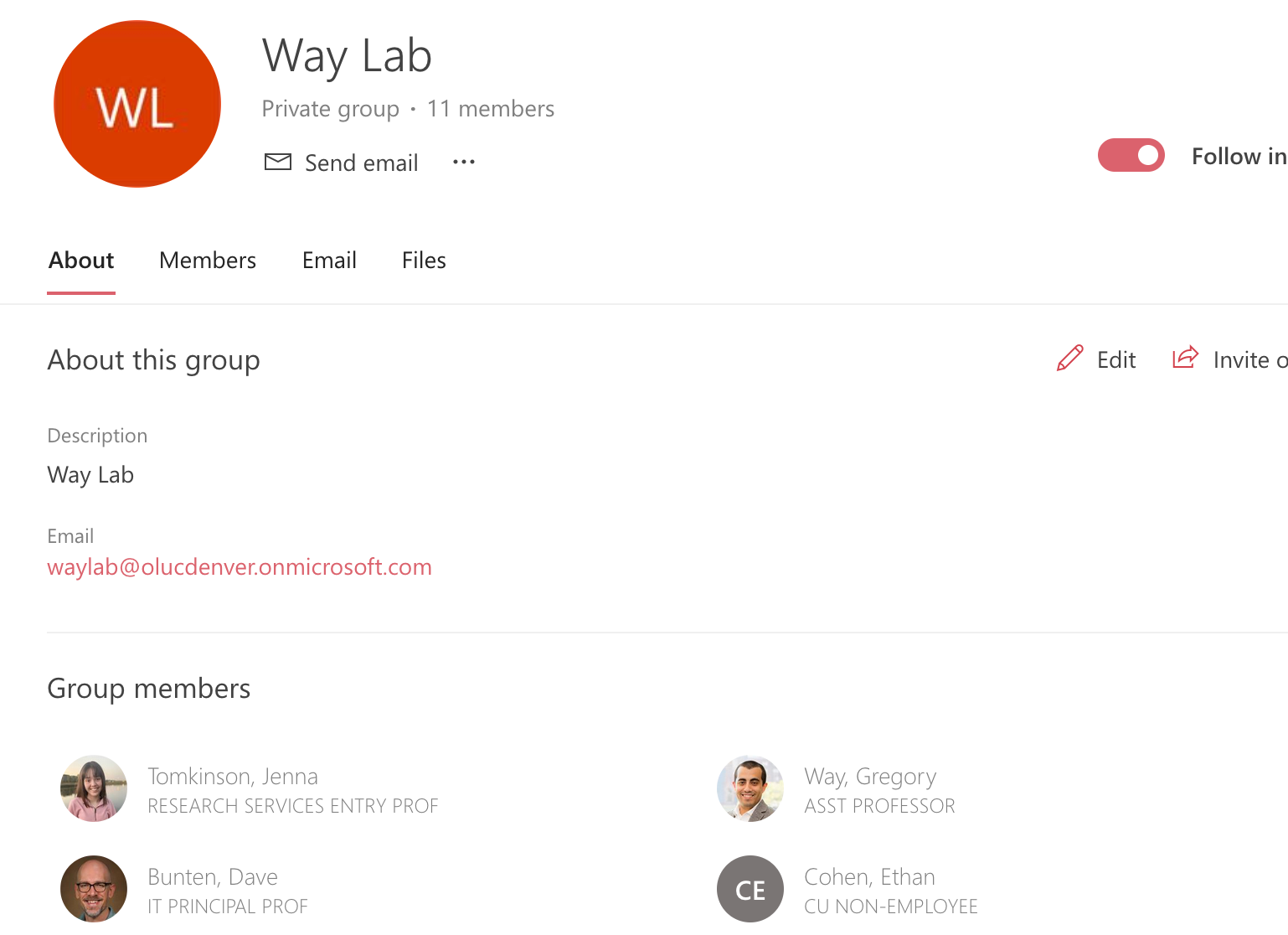
-
From there, you can see all of the members in the group and all emails that have been sent to the group.
@@ -22,10 +23,10 @@ From there, you can see all of the members in the group and all emails that have
## 3) Adding meetings/conferences/vacations/etc to the calendar
-To set up a more transparent environment where every lab member is on the same page, you will need to begin adding events to the Way Lab calendar.
+To set up a more transparent environment where every lab member is on the same page, you will need to begin adding events to the Way Lab calendar.
To do this, you will just create an event normally, but **you will need to make sure that it will be** **created under the Way Lab calendar by having the calendar selected (see above)**.
-You can set what calendar the event is created on using the down arrow button next to the **“Send”** button to select the calendar.
+You can set what calendar the event is created on using the down arrow button next to the **“Send”** button to select the calendar.
In the title of the event, please have the following information in this order:
@@ -41,12 +42,14 @@ You can see if an individual is available or not to meet and change the date and
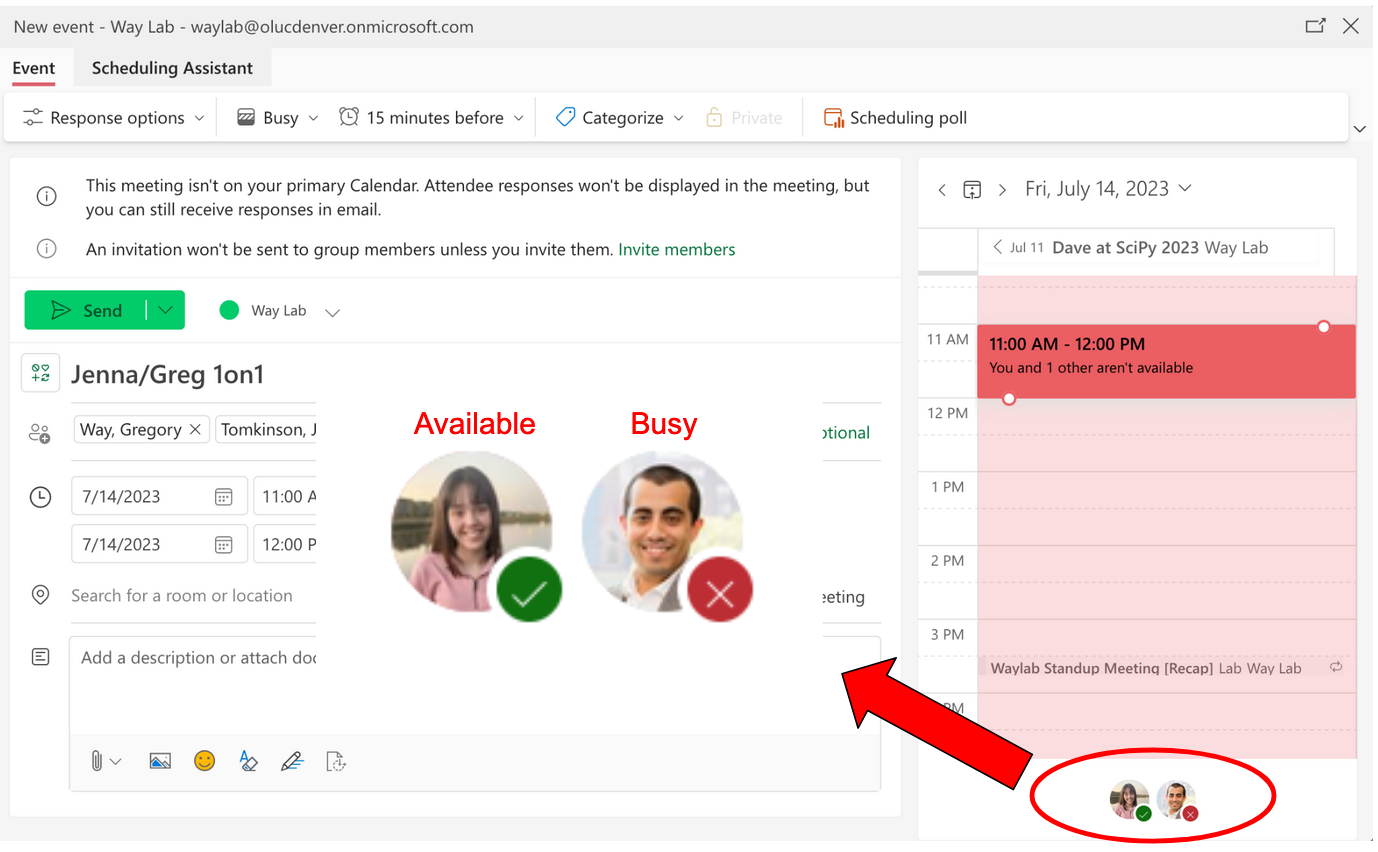
2. For an _appointment or if you will be out_:
+
- Name of individual
- Optional: reason for being out (e.g appointment, class, etc)
- Add “- OOO” to the end
- Example: Jenna Appointment - OOO
-
+
3. For a _vacation or conference_:
+
- Name of individual
- Vacation or name of conference
- Example: Greg SBI2
@@ -57,7 +60,7 @@ For a more organized looking calendar, it is recommended to choose your own colo
For example, one individual’s color is orange and all their events are orange. When you look at the schedule and see an orange event, you know that there is something going on for that individual.
-You can set the color of your event using the top tab called “Categorize”.
+You can set the color of your event using the top tab called “Categorize”.
Make sure to choose a **unique** color that isn’t already being used.
You can check this by viewing the events on the calendar.
diff --git a/internal/computer_setup.md b/internal/computer_setup.md
index a991004..dcac506 100644
--- a/internal/computer_setup.md
+++ b/internal/computer_setup.md
@@ -1,9 +1,10 @@
+# Computer Setup
+
The Way Lab uses Linux distributions for our primary research computing.
Follow these instructions to set up your computer for the first time.
If you’re stuck, ask questions.
All of these steps are likely to change in the future, and, if there are any incomplete or missing details, please modify.
-
## Step 0 - Build your machine!
Chances are you will not need to build your own machine; instead, you will probably have inherited one from a previous lab member.
@@ -12,14 +13,13 @@ In case you are curious, our builds are based off of [these parts](https://pcpar
If you do need to build your machine, you’re not on your own.
You will work with Greg or other lab members to complete the build.
-
## Step 1 - Install the operating system
We are currently experimenting with [Pop!\_OS](https://pop.system76.com/), an open source Ubuntu-based operating system built for STEM.
They’re [located](https://www.google.com/maps/place/System76/@39.7739202,-104.8288135,15z/data=!4m5!3m4!1s0x0:0xf63fca1ea6015d75!8m2!3d39.7739112!4d-104.8288302) just north of Anschutz as well.
Ask Greg, or another lab member, for the live USB.
-The USB contains instructions for your new build to install version 20.04 of Pop!\_OS.
+The USB contains instructions for your new build to install version 20.04 of Pop!\_OS.
**Alternative** - If no live USB is available: Visit this [website](https://pop.system76.com/) and click \`download\`.
We use the latest LTS, and, depending on Step 0, you may install the version with pre-loaded NVIDIA GPU drivers.
@@ -27,7 +27,6 @@ This click will download a \`.iso\` file. Download and open balenaEtcher, and, u
Plug the USB into the new machine.
-
### Step 1.1 - Enter boot mode after powering on
When the motherboard first receives power, it will flash options to enter the boot menu.
@@ -36,13 +35,11 @@ If you’ve successfully entered the menu with the proper USB drive, you will se
Click the proper USB drive, and the operating system will install.
-
### Step 1.1 - Greg will set up using his account
Greg will enter his credentials so that he always has access to all machines.
Don’t worry, we’ll set up your own account soon!
-
## Step 2 - Install core software
Setup github ssh keys
diff --git a/internal/onboarding.md b/internal/onboarding.md
index c52dd0d..63659f0 100644
--- a/internal/onboarding.md
+++ b/internal/onboarding.md
@@ -24,7 +24,7 @@ We will primarily focus on cancer research in our internal projects, and we will
We will establish morphology as a systems biology measurement rivaling molecular readouts in mechanistic information content.
We will understand, with greater detail, how the central dogma of biology links and interacts with other, higher-order biological manifestations, like morphology.
-As research capabilities evolve, new approaches crystalize, and the most pressing problems of tomorrow manifest, we will be ready.
+As research capabilities evolve, new approaches crystallize, and the most pressing problems of tomorrow manifest, we will be ready.
## Expectations
@@ -81,23 +81,28 @@ If at any point you would like to experiment to improve lab policies, please mak
We hold three kinds of regularly scheduled meetings:
1. One-on-ones
- - Once per week.
- - In these meetings, the PI will meet with each individual lab member for one hour and discuss research updates and anything else pertinent to the lab member's success.
- - We will schedule these at the beginning of each semester.
- - We will try to hold all these one-on-one meetings on a single day during core hours.
+
+- Once per week.
+- In these meetings, the PI will meet with each individual lab member for one hour and discuss research updates and anything else pertinent to the lab member's success.
+- We will schedule these at the beginning of each semester.
+- We will try to hold all these one-on-one meetings on a single day during core hours.
+
2. Lab meetings
- - Once per week.
- - We have two different kinds of lab meetings: (1) Journal clubs and (2) Research round tables
- - A journal club presentation consists of either: (A) a deep dive into a single paper, or (B) a quick pass through five different papers extracting out the core message of each.
- - For all journal clubs, the presenter must make the paper's implications to the lab clear.
- - The papers selected must be from a group of papers published since the last presentation.
- - A research round table presentation consists of either: (A) a research in progress, or (B) a white-board brain storming session.
- - For all research round tables, the presenter will lead the discussion and expect lots of questions from other lab members
- - All other lab members not presenting are expected to critically engage with the material
+
+- Once per week.
+- We have two different kinds of lab meetings: (1) Journal clubs and (2) Research round tables
+- A journal club presentation consists of either: (A) a deep dive into a single paper, or (B) a quick pass through five different papers extracting out the core message of each.
+ - For all journal clubs, the presenter must make the paper's implications to the lab clear.
+ - The papers selected must be from a group of papers published since the last presentation.
+- A research round table presentation consists of either: (A) a research in progress, or (B) a white-board brain storming session.
+ - For all research round tables, the presenter will lead the discussion and expect lots of questions from other lab members
+ - All other lab members not presenting are expected to critically engage with the material
+
3. Stand-up
- - Twice per week: Monday and Friday mornings
- - We use scrum for project management and time management.
- - Every member participates and discusses what they are working on this week (or what they accomplished) and if anybody is "blocking" them from their tasks.
+
+- Twice per week: Monday and Friday mornings
+- We use scrum for project management and time management.
+- Every member participates and discusses what they are working on this week (or what they accomplished) and if anybody is "blocking" them from their tasks.
We can also schedule ad-hoc meetings if any lab member needs to practice a presentation, or needs group feedback outside of the normally scheduled meetings.
@@ -172,4 +177,4 @@ We will trust the data we analyze, trust the results of our experiments, and we
### Code of Conduct
We abide by the Code of Conduct as set forth by the CU Anschutz and Center for Health AI communities.
-Please also read our lab's [CODE_OF_CONDUCT.md](CODE_OF_CONDUCT.md) document for more specific details.
\ No newline at end of file
+Please also read our lab's [CODE_OF_CONDUCT.md](../CODE_OF_CONDUCT.md) document for more specific details.
diff --git a/internal/remote_access.md b/internal/remote_access.md
index 421133f..9199dac 100644
--- a/internal/remote_access.md
+++ b/internal/remote_access.md
@@ -1,7 +1,9 @@
+# Remote Access
+
In a hybrid working world, we require solutions to use our powerful computers from anywhere. CU Anschutz OIT office has a solution for us.
Follow these steps to enable ssh or remote desktop access.
-# Secure Socket Shell (SSH)
+### Secure Socket Shell (SSH)
**Step 1A**. On your home computer, install ssh.
@@ -13,8 +15,7 @@ Follow these steps to enable ssh or remote desktop access.
- `brew install openssh`
-
-## Step 2 - Install OpenSSH server on Waylab computer
+### Step 2 - Install OpenSSH server on Waylab computer
**Step 2A**. On your Waylab machine, install openssh-server.
@@ -28,8 +29,7 @@ Follow these steps to enable ssh or remote desktop access.
- Test the install with `sudo service ssh status`
-
-## Step 3 - Connect to Waylab Computer
+### Step 3 - Connect to Waylab Computer
**Step 3A**. Obtain the IP address for your Waylab computer
@@ -45,9 +45,9 @@ Follow these steps to enable ssh or remote desktop access.
- Download Global Protect from CU anschutz website ([link](https://www.ucdenver.edu/offices/office-of-information-technology/software/how-do-i-use/vpn-and-remote-access))
-- Follow the instructions provided CU anschutz (based on Operating System type)
+- Follow the instructions provided CU anschutz (based on Operating System type)
- - [Windows](https://www.ucdenver.edu/docs/default-source/offices-oit-documents/vpn-client-software/windows---globalprotect-vpn-installation-instructions.pdf?sfvrsn=e1579eb8_6)
+ - [Windows](https://www.ucdenver.edu/docs/default-source/offices-oit-documents/vpn-client-software/windows---globalprotect-vpn-installation-instructions.pdf?sfvrsn=e1579eb8_6)
- [MacOs](https://www.ucdenver.edu/docs/default-source/offices-oit-documents/vpn-client-software/mac---globalprotect-vpn-installation-instructions.pdf?sfvrsn=89559eb8_11)
@@ -55,9 +55,9 @@ Follow these steps to enable ssh or remote desktop access.
- Once download it, open up Global Protect VPN
- - Enter CU anschutz username and password (this step assumes you have already follow the installation instructions)
+ - Enter CU anschutz username and password (this step assumes you have already follow the installation instructions)
- - Once you see the \`connected\` message, your VPN is active!
+ - Once you see the \`connected\` message, your VPN is active!
**Step 3D**. Login using your Waylab computer username on your home computer
@@ -65,26 +65,23 @@ Follow these steps to enable ssh or remote desktop access.
- If you are connecting for the first time you will need to hit ENTER to add the appropriate ECDSA key fingerprint
-- You should see the name of the remote computer in your terminal.
+- You should see the name of the remote computer in your terminal.
For more details see this post:
+## Remote Desktop
-# Remote Desktop
-
-## Step 1 - Is remote desktop enabled?
+### Step 1 - Is remote desktop enabled?
**Step 1A**. Check [waylab computers](https://docs.google.com/spreadsheets/d/1xTQ9eLwazsbBByJ0mxU5D9X4t-sJlr2m3k_sp2nrBr4/edit#gid=0) to see if remote desktop is enabled.
**Step 1B**. If not, reach out to OIT office by filing a ticket (go to the [OIT portal](https://servicecenter.oit.ucdenver.edu/CherwellPortal/IT?_=67e6572e#0) and submit a ticket include details of computer name and IP address in the ticket)
-
-## Step 2 - Download xRDP on your Waylab Machine
+### Step 2 - Download xRDP on your Waylab Machine
**Step 2A**. Follow these instructions:
-
-## Step 3 - Download vmware horizon on your home computer
+### Step 3 - Download vmware horizon on your home computer
**Step 3A**. Navigate to
@@ -94,8 +91,7 @@ For more details see this post: